No mod ratios in csv file #62
-
|
Hello, I am running the program on my data set, and it is able to generate the csv file but in the modification columns it only outputs a 0. Am I doing some wrong in my data prep upstream of the analysis? I start by indexing my gzipped fast5 files with the fastq files using nanopolish index, then I align using eventalign followed by the data prep and mod inference modules of m6anet. Thanks! |
Beta Was this translation helpful? Give feedback.
Replies: 2 comments 2 replies
-
|
hi @esh97 Can you show me the first 5 rows of your nanopolish eventalign.txt and your result files? Thanks! |
Beta Was this translation helpful? Give feedback.
-
|
Hi @esh97 , your eventalign file does not have the start_index and end_index column that tells m6anet how to segment your reads. When running nanopolish make sure you include the -signal-index flag such as: nanopolish eventalign –reads reads.fastq –bam reads.sorted.bam –genome transcript.fa –scale-events –signal-index –summary |
Beta Was this translation helpful? Give feedback.
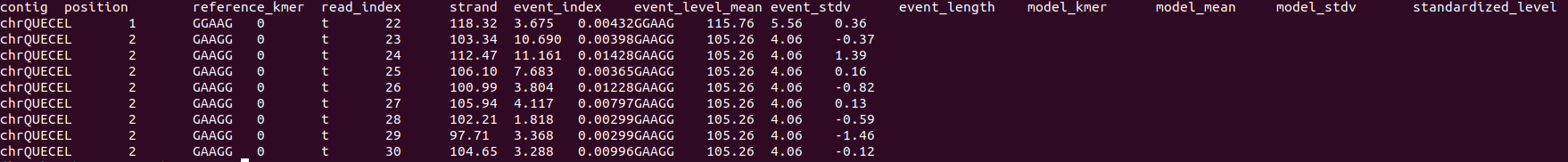
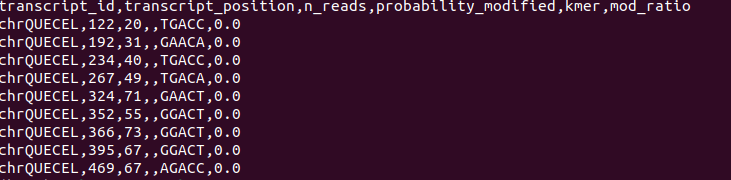
Hi @esh97 , your eventalign file does not have the start_index and end_index column that tells m6anet how to segment your reads. When running nanopolish make sure you include the -signal-index flag such as:
nanopolish eventalign –reads reads.fastq –bam reads.sorted.bam –genome transcript.fa –scale-events –signal-index –summary
/path/to/summary.txt –threads 50 > /path/to/eventalign.txt