About addGeneIntegrationMatrix #1023
Unanswered
tzhu-bio
asked this question in
Questions / Documentation
Replies: 2 comments 11 replies
-
|
This behavior is not observed using the tutorial data on my end. |
Beta Was this translation helpful? Give feedback.
5 replies
-
|
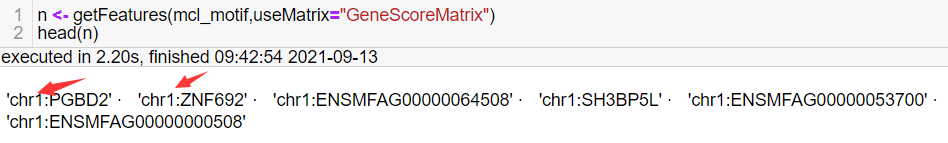
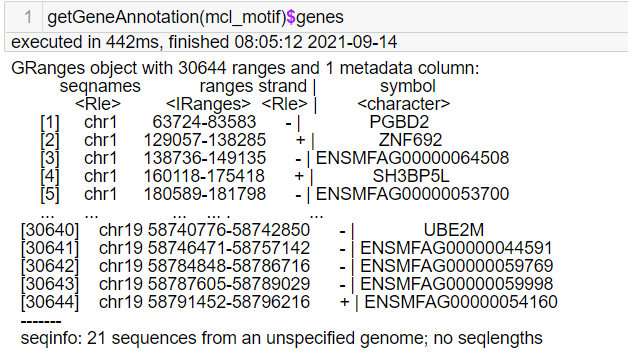
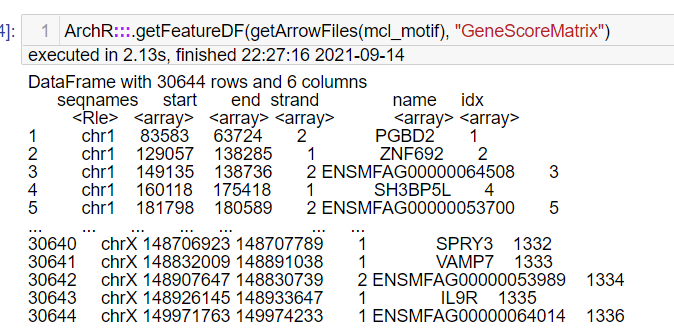
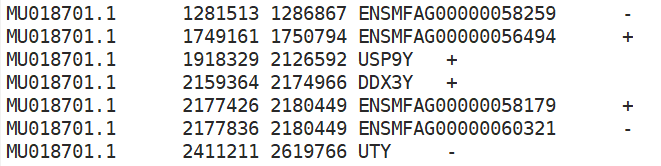
Sorry to bother you again. I got the features from the GeneScore matrix and why they all start with "chr". This is not the same as the tutorial. I don't think I made a mistake in constructing the custom genome. |
Beta Was this translation helpful? Give feedback.
6 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment

Uh oh!
There was an error while loading. Please reload this page.
-
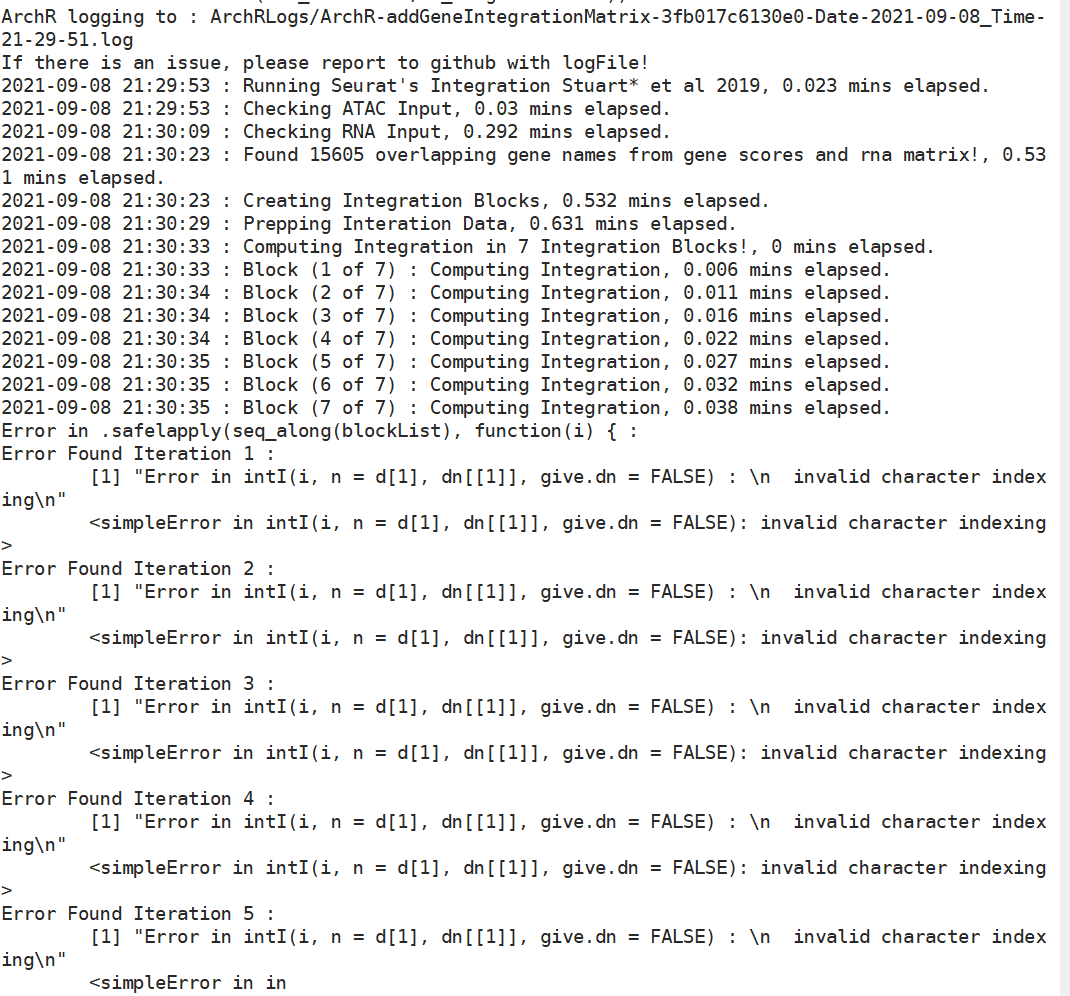
When I used addGeneIntegrationMatrix to integrate the ATAC and RNA, it only found 15605 overlapping gene names from gene scores and rna matrix! Is this a relatively few overlapping genes?
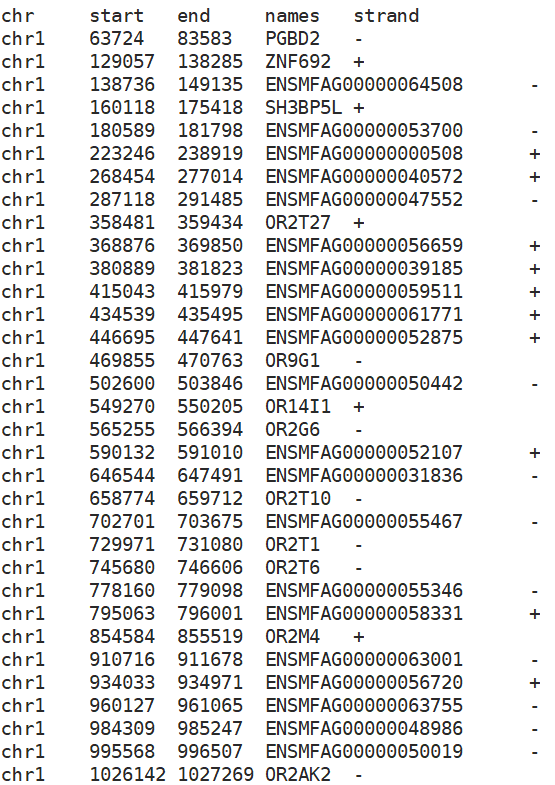
And I found the GeneIntegrationMatrix features only has the "ENSMFAGxxxxxxxx" gene id but with no gene name like "PGBD2"....
Then I change the rna matrix gene name, which are the same as genescore matrix gene name, but the result did not change.
Beta Was this translation helpful? Give feedback.
All reactions