interpreting motif deviation scores #1109
Unanswered
mitchTwoTimes
asked this question in
Questions / Documentation
Replies: 1 comment
-
|
yes |
Beta Was this translation helpful? Give feedback.
0 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
Hello,
Can you please help me understand if I'm interpreting the motif deviations correctly? I found these TFs through "positive regulator" analysis using the gene expression matrix as the correlate to my motif matrix.
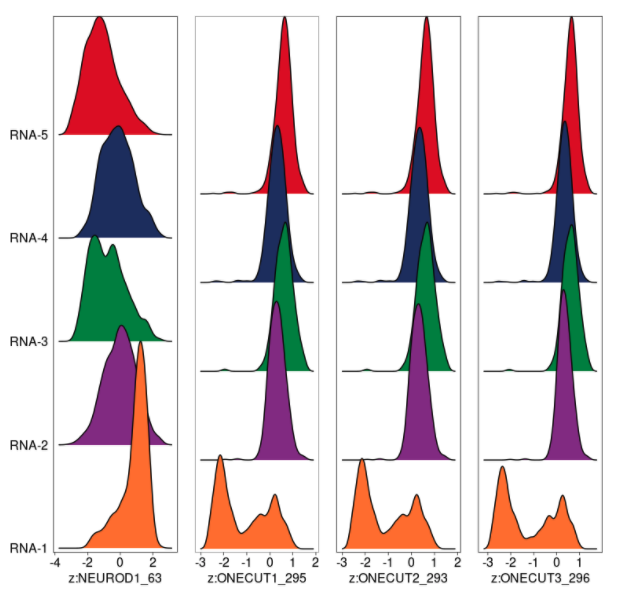
Let me focus on NEUROD1. To me, this data says the genome-wide motifs that NEUROD1 is expected to bind to are more accessible in "RNA-1" relative to the other clusters. Is that accurate?
Thanks
Beta Was this translation helpful? Give feedback.
All reactions