UMAP cluster separation with bins vs. peaks #1116
-
|
Hi @rcorces, I compared 500 bp bins (estimated LSI: iterations=2, first selection= 'top', varfeatures=50,000, dims=1:30) and Signac top features (q0 - all the features) after calling peaks with macs2. I noticed that even after tweaking several parameters such as increasing the iterations or decreasing the varfeatures, I get more separated clusters using peaks than bins. Here are the plots (first one bins and second peaks - in both cluster resolution=0.3). I really like the idea of using bins and understand that separation between UMAP clusters is more technical than biological, however, I am wondering if you can suggest which parameters to tweak to get more separated clusters? In addition, I am having a bit of difficulty in understanding the use of the parameters provided in LSI. Thank you very much!! |
Beta Was this translation helpful? Give feedback.
Replies: 2 comments 5 replies
-
|
To clarify, clusters are called in the LSI space, regardless of how they are plotted in UMAP space. You can also force |
Beta Was this translation helpful? Give feedback.
-
|
Hi @AnjaliC4! I have had similar observations with our data, I was wondering if you figured out how to get more separated clusters like with using Signac? |
Beta Was this translation helpful? Give feedback.
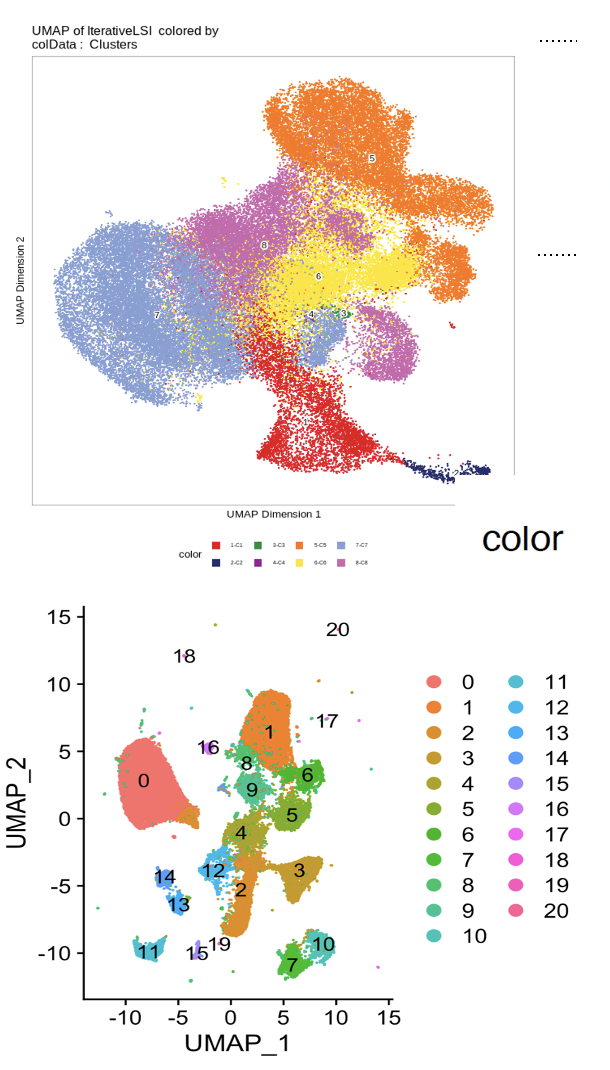
To clarify, clusters are called in the LSI space, regardless of how they are plotted in UMAP space.
I think you would have better luck tweaking the parameters of the UMAP if you are looking to get more separation. for example
minDistYou can also force
addIterativeLSI()to mimic your results from Signac by tweaking the parameters. In our hands, using defined metrics for assessing clustering (i.e. not subjective measures like how separate the clusters are), ArchR performs very well.