Version of ArchR hg38 genome? #1658
Unanswered
RegnerM2015
asked this question in
Questions / Documentation
Replies: 1 comment
-
|
Your UCSC genome browser is showing Gencode genes. Those are quite a bit different from the genes described by the TxDB object |
Beta Was this translation helpful? Give feedback.
0 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
Hi @rcorces,
Would you be able to share what specific version of h38 precompiled ArchR uses?
I see that this is made from BSgenome.Hsapiens.UCSC.hg38, TxDb.Hsapiens.UCSC.hg38.knownGene, org.Hs.eg.db according to documentation.
I ask because there is a lncRNA missing from this precompiled annotation compared to the current UCSC genome browser.
ArchR gene track (using hg38 precompiled default) at chr2:118797677−118857678:
UCSC track at chr2:118797677−118857678:
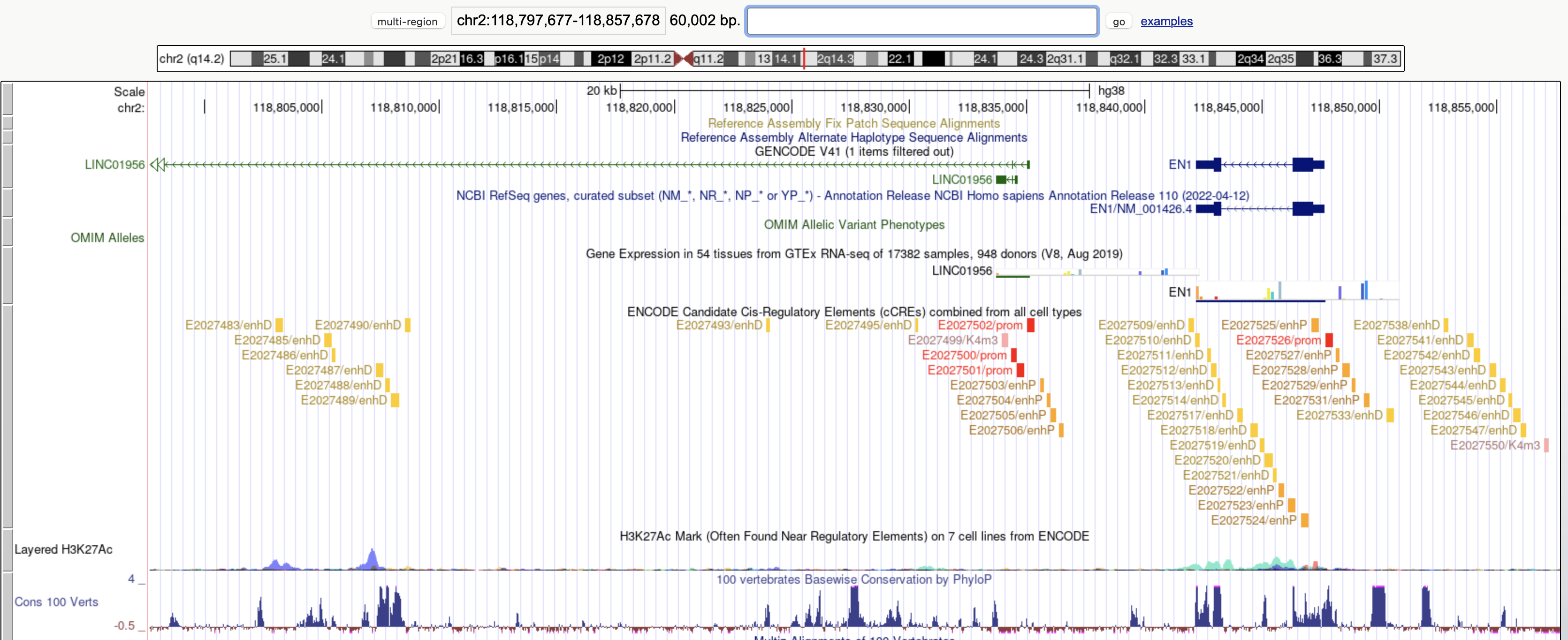
I think the discrepancy here between ArchR and UCSC may be caused by a difference in the annotation set and/or version (UCSC perhaps showing a newer version which happens to add in this lncRNA LINC01956). Thus, distal peaks in this region (left of EN1) would presumably become intronic if this lncRNA LINC01956 was included in the annotation.
Thanks for your help!
Beta Was this translation helpful? Give feedback.
All reactions