How is "nearest gene" determined? #1675
-
Beta Was this translation helpful? Give feedback.
Replies: 1 comment 4 replies
-
|
I think you've gone a little too far into searching for something. What you're looking at is the metadata of the object and specifically the params that were used when calling The nearest gene is not stored in the object explicitly. You would have to match the values in the |
Beta Was this translation helpful? Give feedback.
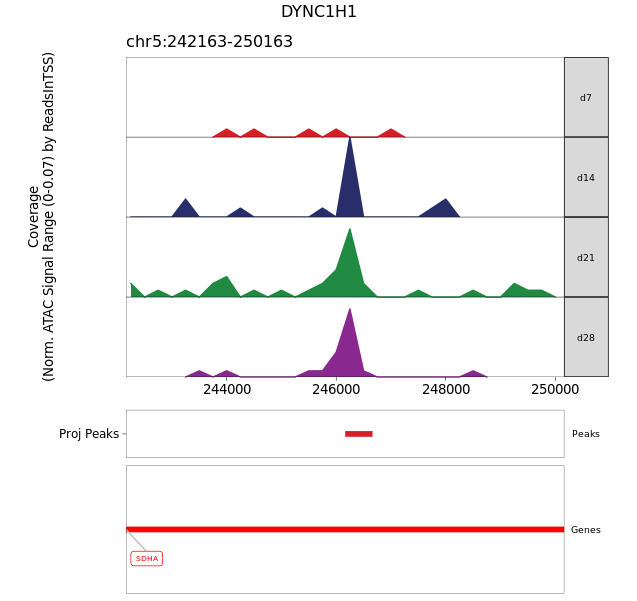
I think you've gone a little too far into searching for something. What you're looking at is the metadata of the object and specifically the params that were used when calling
getMarkerFeatures(). One of those params was theArchRProjectobject. By looking atmarkerTest@metadata$Params$ArchRProj@peakSet$nearestGene, you are effectively just doingArchRProj@peakSet$nearestGenewhich is actually disconnected from the analysis.The nearest gene is not stored in the object explicitly. You would have to match the values in the
idxcolumn of theSummarizedExperimentwith the values i…