Replies: 2 comments 4 replies
-
|
I'm not sure I understand your issue. Perhaps you can read this previous post and clarify. |
Beta Was this translation helpful? Give feedback.
0 replies
-
Beta Was this translation helpful? Give feedback.
4 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
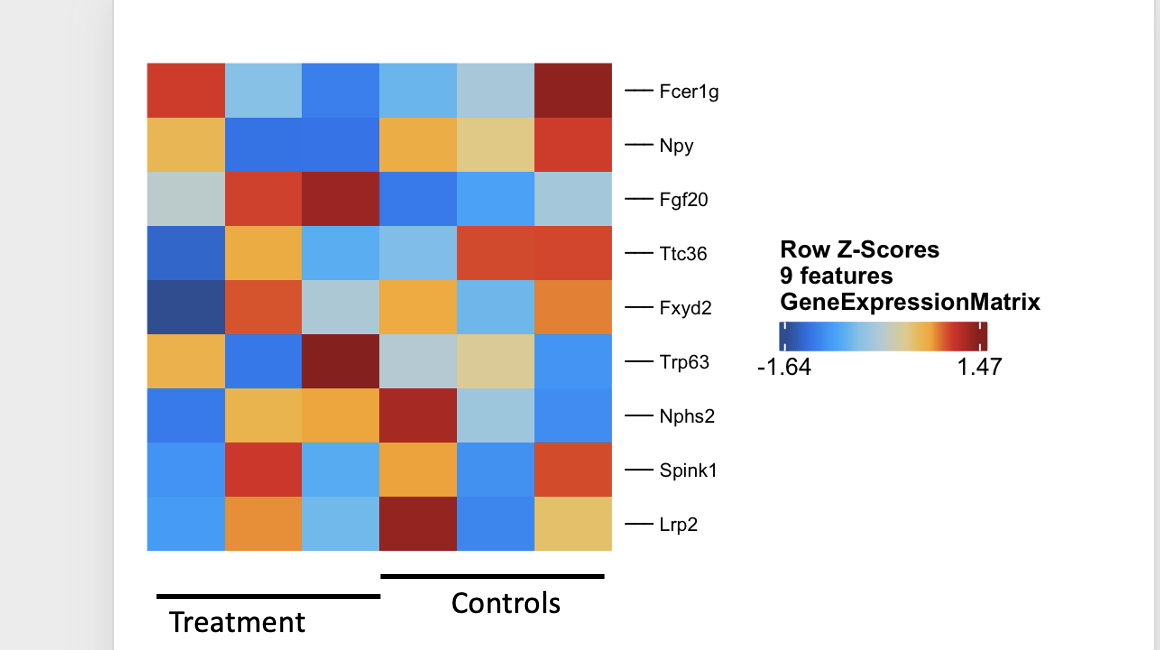
@rcorces
Hi Ryan,
When running getMarkerFeatures(proj) on replicates for a given treatment I obtain big differences when querying for specific markers using plotMarkerHeatmap(). This is confounding the inference from different treatments.
se <- getMarkerFeatures(ArchRProj = proj, groupBy = "Sample", useMatrix = "GeneExpressionMatrix", ### bias =c("TSSEnrichment", "log10(Gex_nUMI)"))
Outside of bias adjustments what can be done to address this issue? Can you help with some suggestions.
Thank you!
Beta Was this translation helpful? Give feedback.
All reactions