Questions about ArchR synthetic doublet generation #594
Replies: 1 comment 5 replies
-
Sometimes but not always - I think you would be surprised how unbalanced doublets can be.
Yes - we probably could be more clear on this in the paper and manual. Homo-typic doublets are not captured but I havent seen any approach that doesnt use genotype able to accurately capture homotypic doublets. We show the performance of just using the number of fragments in the manuscript and it is not very effective.
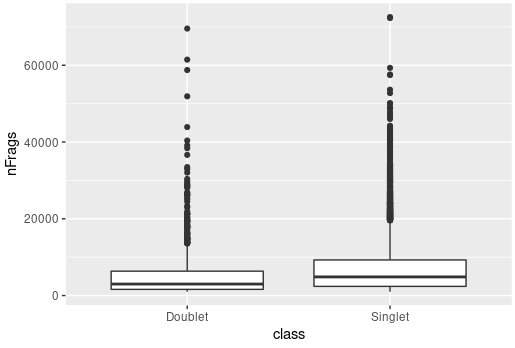
You're welcome to benchmark this yourself on our cell line data but I think you'll find your hypothesis to not play out as expected. A cursory check shows this though I did this very quickly: |
Beta Was this translation helpful? Give feedback.
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
I love the ideas behind archR and scrublet doublet predictions - very interesting and creative. I would like to know more about how the synthetic doublets are generated.
In section 2.1 of the ArchR manual it says "To predict which “cells” are actually doublets, we synthesize in silico doublets from the data by mixing the reads from thousands of combinations of individual cells." In the associated figure, it shows groups of 2 cells being combined and then divided by two
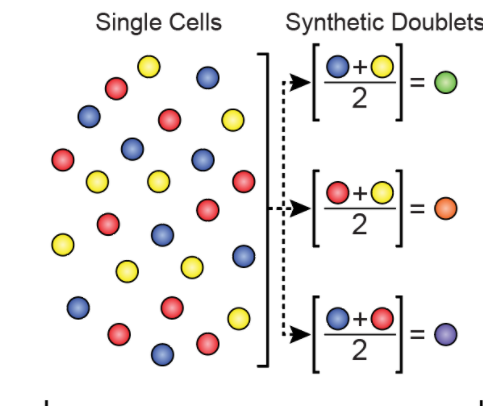
First question, are all synthetic doublets divided by two, as shown in the figure? Wouldn't a doublet contain the sum of fragments from two cells rather than dividing them by two? Also, wouldn't this method not be able to properly identify doublets of the same cell type? Example: a hybrid of two astrocytes would still look like an astrocyte, leading to false positives. For this reason, I believe a higher fragment count should also be accounted for in doublet generation.
In the nature genetics paper: "To validate this approach, we carried out scATAC-seq on a mixture of ten human cell lines (n = 38,072 cells), allowing for genotype-based identification of doublets via demuxlet as a ground-truth comparison for computational identification of doublets by ArchR". This only identifies doublets based on genotype - What about cells that failed to dissociate properly in nuclei isolation (i.e. same donor, same genotype)? Depending on the tissue composition, cells that didn't dissociate could often be the same cell type, leading me back to my first question.
I feel like a solution to these problems could be to generate two synthetic doublets from each cell-cell pair: the first using your original method, and the second accounting for the higher read count of doublets (i.e. sum of the fragments)
Beta Was this translation helpful? Give feedback.
All reactions