How are peaks annotated as intronic/exonic/etc? #687
Unanswered
Dragonlongzhilin
asked this question in
Questions / Documentation
Replies: 1 comment
-
|
Please note that questions and feature requests should no longer be submitted as issues. I have migrated your issue to a Discussion topic which is the appropriate place. Only bugs should be reported under issues. This is indicated in the Issue Template which we would appreciate if all users would utilize. Peak annotations are determined based on your gene annotation. Presumably there is at least one isoform of a gene (not necessarily CA9) in your gene annotation where there is an exon that at least partially overlaps that peak. In this case CA9 is the gene with the closest annotated start site. |
Beta Was this translation helpful? Give feedback.
0 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
I want to view the peak of CA9 gene.
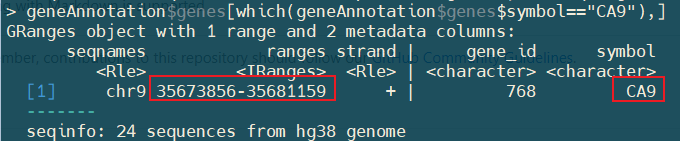
The genome position of this gene is chr9: 35673856-35681159
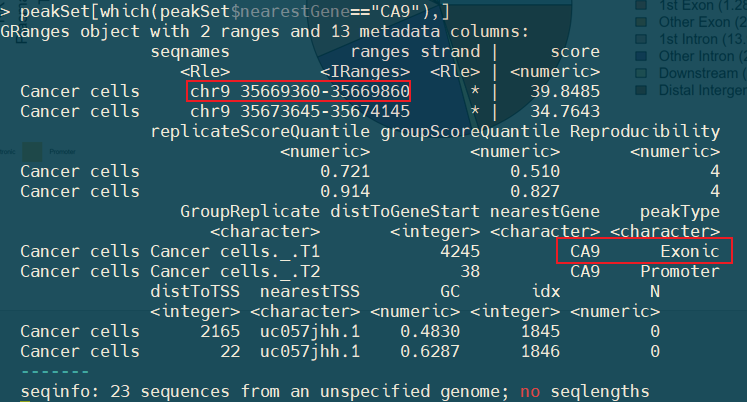
I found that two peaks fall into this gene and extracted the result from PeakSet matrix.
But there was a peak (chr9:35669360-35669860) annotated as Exonic, but its genome location is upstream of this gene.
I did't kown why?
Beta Was this translation helpful? Give feedback.
All reactions