About data integration #827
Unanswered
tzhu-bio
asked this question in
Questions / Documentation
Replies: 1 comment 2 replies
-
|
Thanks for using ArchR. I'm afraid that your question doesnt really have an answer. snATAC-seq data from different samples can be very effectively integrated, which we show in our publication combining datasets generated across multiple different platforms (Fluidigm, biorad, 10x Genomics) etc. If you are having trouble, perhaps your data is highly confounded by batch effects or you need to tune the parameters of your dimensionality reduction / try using Harmony. |
Beta Was this translation helpful? Give feedback.
2 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
-
Thanks for your ArchR!
Why is it that using ArchR to integrate scATAC data does not integrate well and each sample is separated from each other?
The seurat integration of RNA data allows for good integration between different samples.
Is this the cause of the data type?
ATAC(using ArchR)
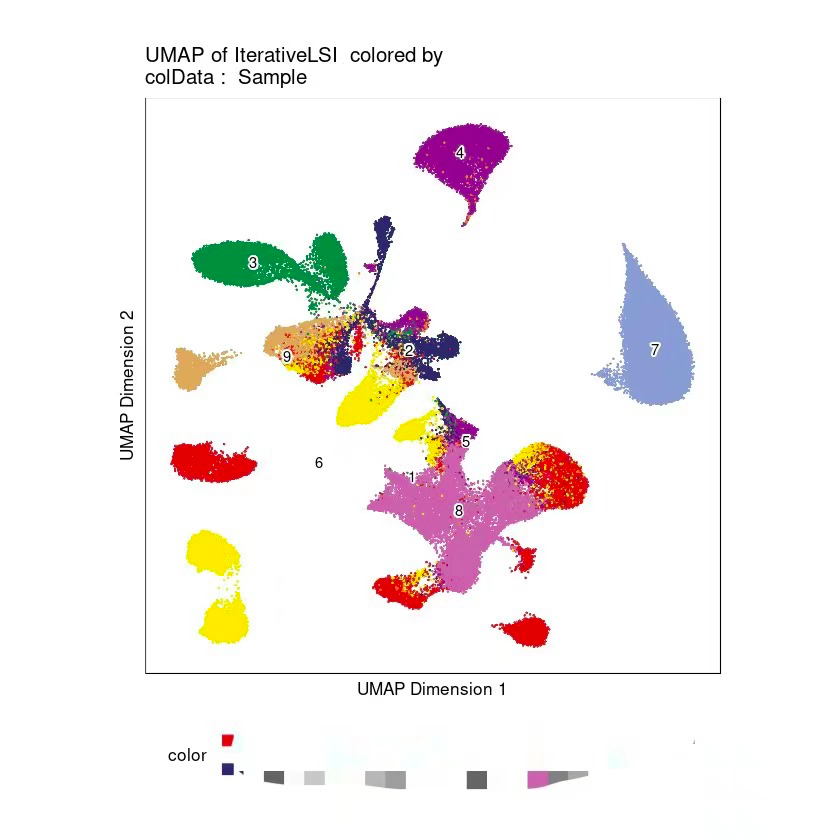
RNA(using Seurat)
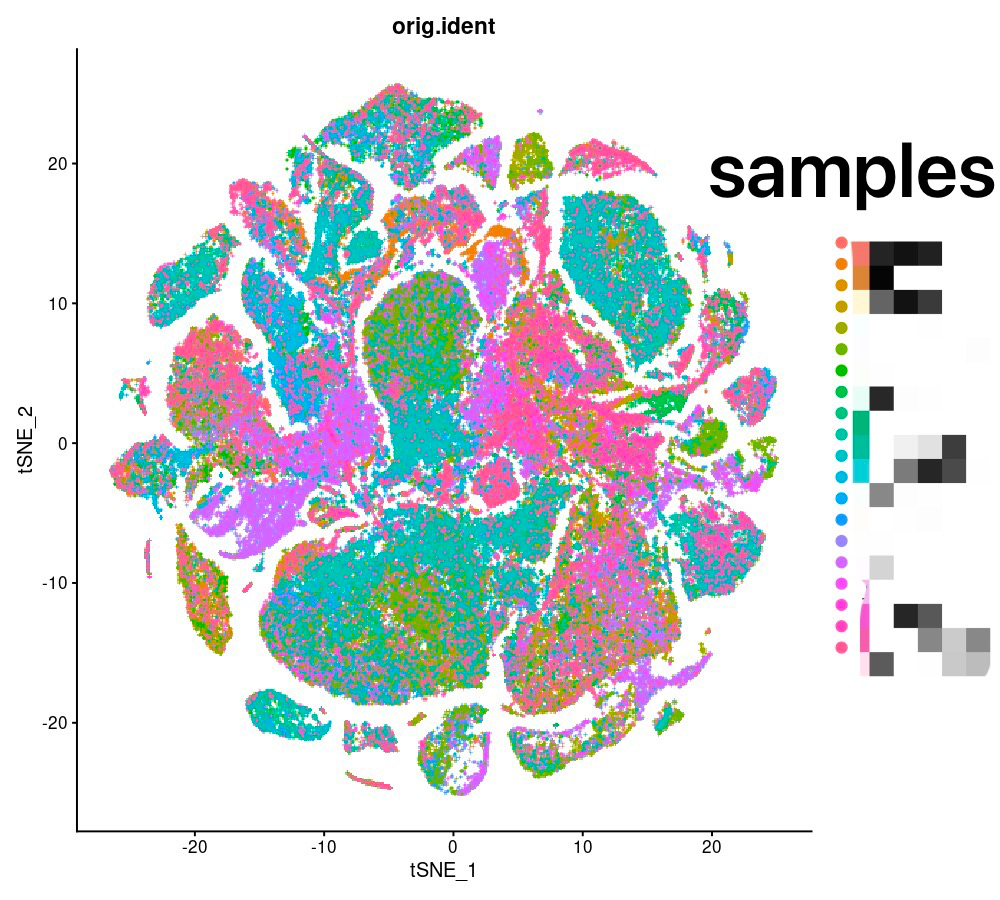
Beta Was this translation helpful? Give feedback.
All reactions