Harmony correction results in no UMAP separation #994
Unanswered
RegnerM2015
asked this question in
Questions / Documentation
Replies: 1 comment 5 replies
-
|
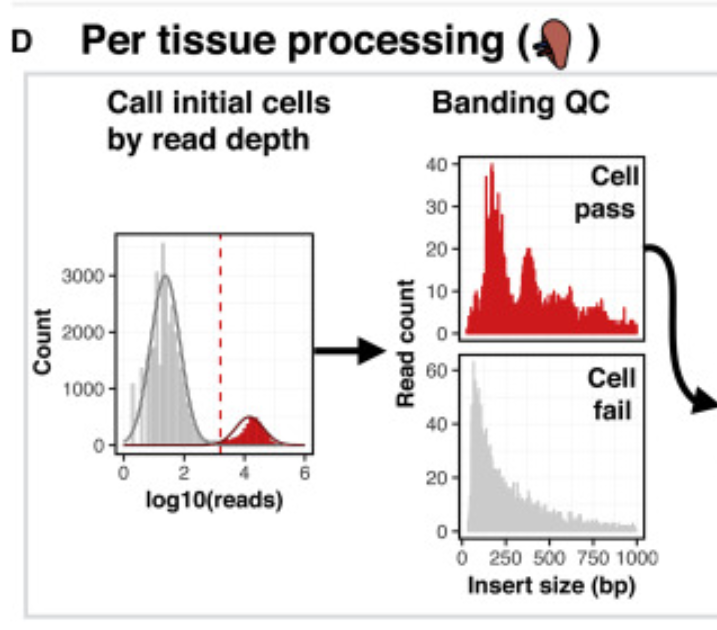
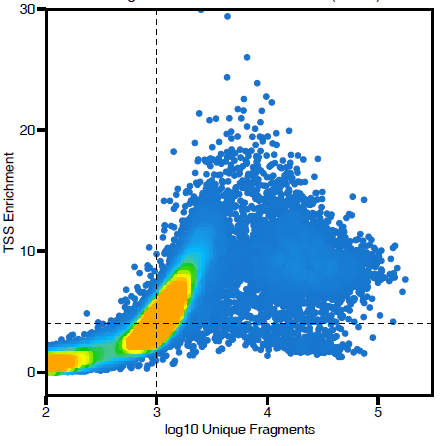
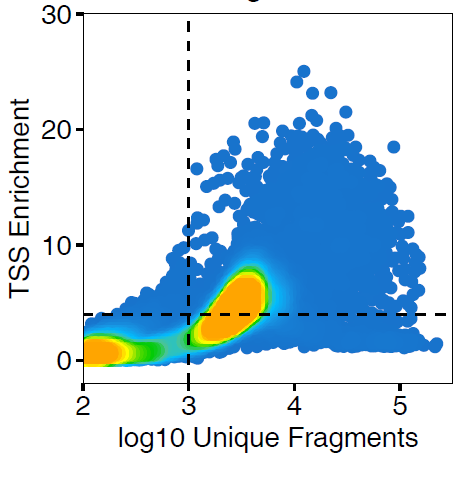
There is no right answer here. I would try all of the things that you listed. In terms of QC, just make sure that you are inspecting the reads x TSS QC plots and selecting cutoffs that make sense. In my opinion, its ok to use a different cutoff for number of frags for different samples in order to exclude junk (but I would keep the TSS cutoff the same whenever possible). |
Beta Was this translation helpful? Give feedback.
5 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
-
Hi @rcorces and @jgranja24,
Please note that I am using ArchR version 1.0.1
I have 17 samples profiled with 10x Genomics scATAC-seq. To mitigate batch/patient of origin effects, I decided to run Harmony after adding the LSI embeddings. There are 122,493 cells after quality control (minTSS = 6, minFrags = 1000, maxFrags =1e+05, doublets removed using filterDoublets() with default parameters). These seem like reasonable quality control cutoffs, but when I visualize the Harmony-based UMAP, I observe a large blob of cells with little separation. Please see the UMAP plots attached, highlighted by sample, by TSSEnrichment, and by log10(nFrags).
Which seems to be the most reasonable path forward?
Below are the PDF files and code used to generate these plots:
Plot-UMAP-TSS.pdf
Plot-UMAP-Sample.pdf
Plot-UMAP-logFrags.pdf
Beta Was this translation helpful? Give feedback.
All reactions