|
| 1 | +# ITKColorNormalization |
| 2 | + |
| 3 | + |
| 4 | + |
| 5 | +[  ](https://pypi.python.org/pypi/itk-spcn) |
| 6 | + |
| 7 | + |
| 8 | + |
| 9 | +## Overview |
| 10 | + |
| 11 | +This [Insight Toolkit (ITK)](https://itk.org/) module performs Structure Preserving Color Normalization on an [H & |
| 12 | +E](https://en.wikipedia.org/wiki/H%26E_stain) image using a reference image. The module is in C++ and is also packaged |
| 13 | +for [Python](https://www.python.org/). |
| 14 | + |
| 15 | +H & E ([hematoxylin](https://en.wikipedia.org/wiki/Haematoxylin) and [eosin](https://en.wikipedia.org/wiki/Eosin)) are |
| 16 | +stains used to color parts of cells in a histological image, often for medical diagnosis. Hematoxylin is a compound |
| 17 | +that stains cell nuclei a purple-blue color. Eosin is a compound that stains extracellular matrix and cytoplasm pink. |
| 18 | +However, the exact color of purple-blue or pink can vary from image to image, and this can make comparison of images |
| 19 | +difficult. This routine addresses the issue by re-coloring one image (the first image supplied to the routine) using |
| 20 | +the color scheme of a reference image (the second image supplied to the routine). The technique requires that the |
| 21 | +images have at least 3 colors, such as red, green, and blue (RGB). |
| 22 | + |
| 23 | +Structure Preserving Color Normalization is a technique described in [Vahadane et al., |
| 24 | +2016](https://doi.org/10.1109/TMI.2016.2529665) and modified in [Ramakrishnan et al., |
| 25 | +2019](https://arxiv.org/abs/1901.03088). The idea is to model the color of an image pixel as something close to pure |
| 26 | +white, which is reduced in intensity in a color-specific way via an optical absorption model that depends upon the |
| 27 | +amounts of hematoxylin and eosin that are present. [Non-negative matrix |
| 28 | +factorization](https://en.wikipedia.org/wiki/Non-negative_matrix_factorization) is used on each analyzed image to |
| 29 | +simultaneously derive the amount of hematoxylin and eosin stain at each pixel and the image-wide effective colors of |
| 30 | +each stain. |
| 31 | + |
| 32 | +The implementation in ITK accelerates the non-negative matrix factorization by choosing the initial estimate for the |
| 33 | +color absorption characteristics using a technique mimicking that presented in [Arora et al., |
| 34 | +2013](http://proceedings.mlr.press/v28/arora13.html) and modified in [Newberg et al., |
| 35 | +2018](https://doi.org/10.1371/journal.pone.0193067). This approach finds a good solution for a non-negative matrix |
| 36 | +factorization by first transforming it to the problem of finding a convex hull for a set of points in a cloud. |
| 37 | + |
| 38 | +The lead developer of InsightSoftwareConsortium/ITKColorNormalization is [Lee Newberg](https://github.com/Leengit/). |
| 39 | + |
| 40 | +## Installation for Python |
| 41 | + |
| 42 | +ITKColorNormalization and all its dependencies can be easily installed with [Python |
| 43 | +wheels](https://blog.kitware.com/itk-is-on-pypi-pip-install-itk-is-here/). Wheels have been generated for macOS, Linux, |
| 44 | +and Windows and several versions of Python, 3.5, 3.6, 3.7, and 3.8. If you do not want the installation to be to your |
| 45 | +current Python environment, you should first create and activate a [Python virtual environment |
| 46 | +(venv)](https://docs.python.org/3/tutorial/venv.html) to work in. Then, run the following from the command-line: |
| 47 | + |
| 48 | +```shell-script |
| 49 | +pip install itk-spcn |
| 50 | +``` |
| 51 | + |
| 52 | +Launch `python`, import the itk package, and set variable names for the input images |
| 53 | + |
| 54 | +```python |
| 55 | +import itk |
| 56 | +input_image_filename = 'path/to/image_to_be_normalized' |
| 57 | +reference_image_filename = 'path/to/image_to_be_used_as_color_reference' |
| 58 | +``` |
| 59 | + |
| 60 | +## Usage in Python |
| 61 | + |
| 62 | +The following example transforms this input image |
| 63 | + |
| 64 | +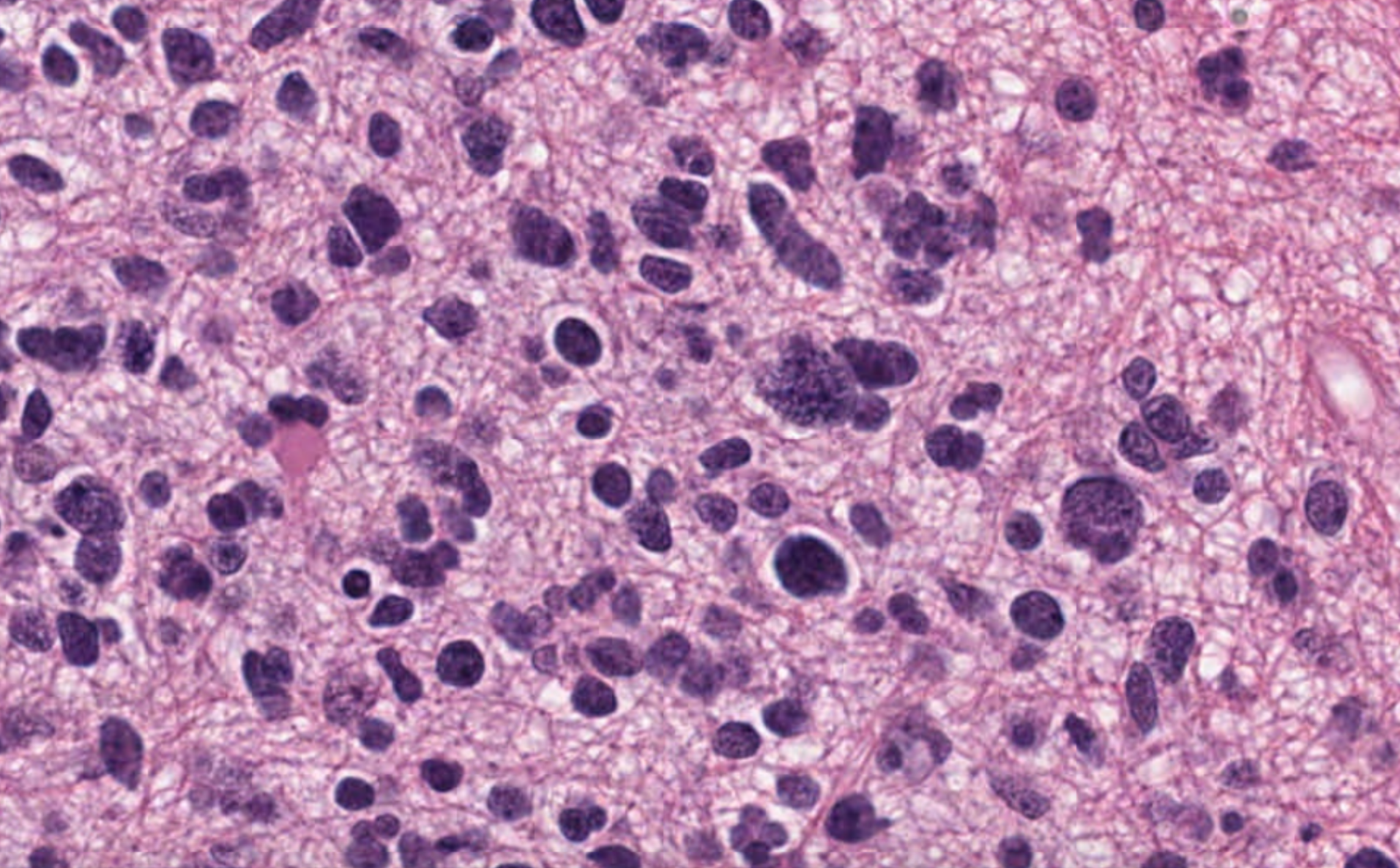 |
| 65 | + |
| 66 | +using the color scheme of this reference image |
| 67 | + |
| 68 | +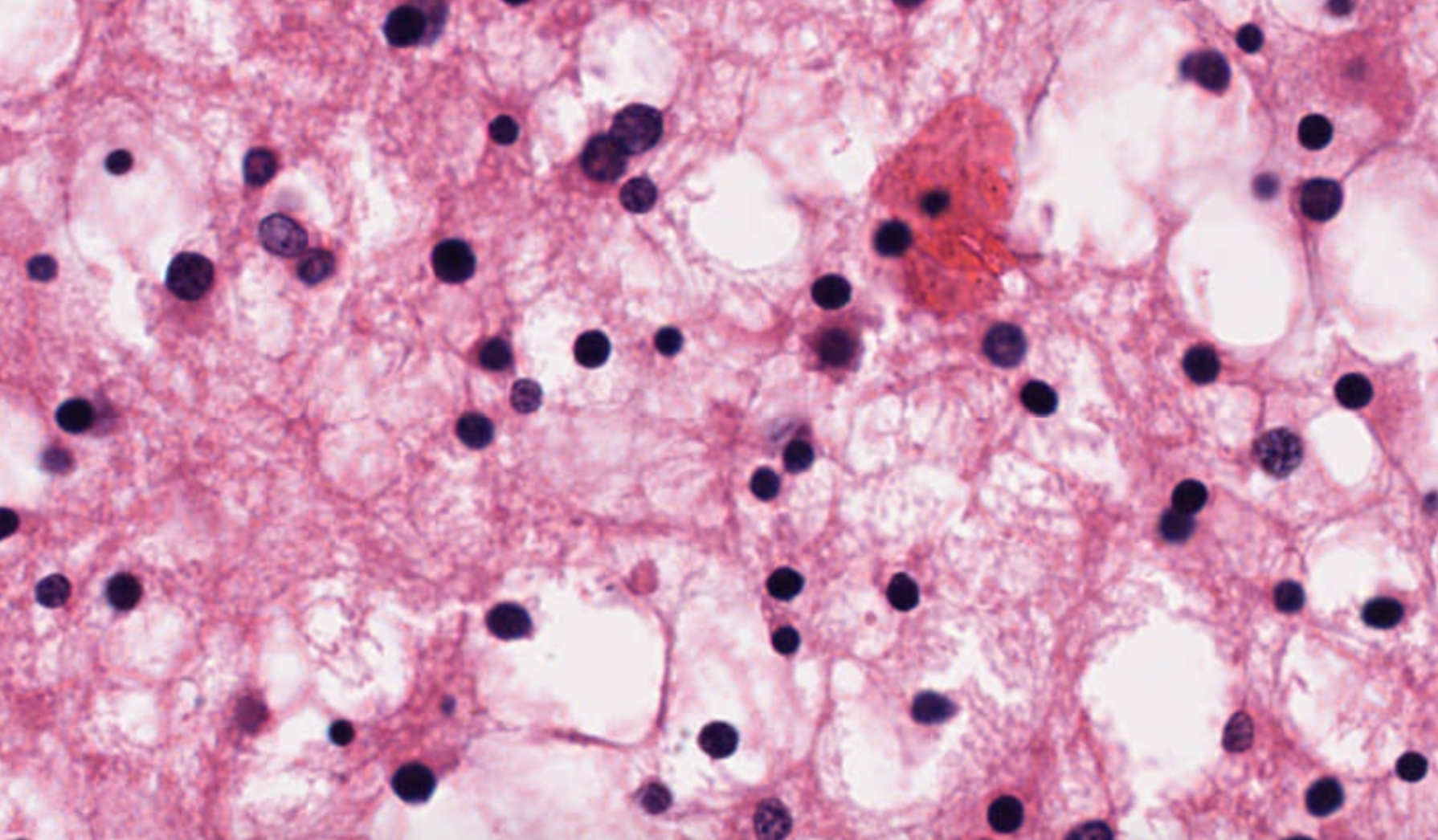 |
| 69 | + |
| 70 | +to produce this output image |
| 71 | + |
| 72 | + |
| 73 | + |
| 74 | +### Functional interface to ITK |
| 75 | + |
| 76 | +You can use the functional, eager interface to ITK to choose when each step will be executed as follows. The |
| 77 | +`input_image` and `reference_image` are processed to produce `normalized_image`, which is the `input_image` with the |
| 78 | +color scheme of the `reference_image`. The `color_index_suppressed_by_hematoxylin` and |
| 79 | +`color_index_suppressed_by_eosin` arguments are optional if the `input_image` pixel type is RGB or RGBA. Here you are |
| 80 | +indicating that the color channel most suppressed by hematoxylin is 0 (which is red for RGB and RGBA pixels) and that |
| 81 | +the color most suppressed by eosin is 1 (which is green for RGB and RGBA pixels)\; these are the defaults for RGB and |
| 82 | +RGBA pixels. |
| 83 | + |
| 84 | +```python |
| 85 | +input_image = itk.imread(input_image_filename) |
| 86 | +reference_image = itk.imread(reference_image_filename) |
| 87 | + |
| 88 | +eager_normalized_image = itk.structure_preserving_color_normalization_filter( |
| 89 | + input_image, |
| 90 | + reference_image, |
| 91 | + color_index_suppressed_by_hematoxylin=0, |
| 92 | + color_index_suppressed_by_eosin=1) |
| 93 | + |
| 94 | +itk.imwrite(eager_normalized_image, output_image_filename) |
| 95 | +``` |
| 96 | + |
| 97 | +### ITK pipeline interface |
| 98 | + |
| 99 | +Alternatively, you can use the ITK pipeline infrastructure that waits until a call to `Update()` or `Write()` before |
| 100 | +executing the pipeline. The function `itk.StructurePreservingColorNormalizationFilter.New()` uses its argument to |
| 101 | +determine the pixel type for the filter\; the actual image is not used there but is supplied with the |
| 102 | +`spcn_filter.SetInput(0, input_reader.GetOutput())` call. As above, the calls to |
| 103 | +`SetColorIndexSuppressedByHematoxylin` and `SetColorIndexSuppressedByEosin` are optional if the pixel type is RGB or |
| 104 | +RGBA. |
| 105 | + |
| 106 | +```python |
| 107 | +input_reader = itk.ImageFileReader.New(FileName=input_image_filename) |
| 108 | +reference_reader = itk.ImageFileReader.New(FileName=reference_image_filename) |
| 109 | + |
| 110 | +spcn_filter = itk.StructurePreservingColorNormalizationFilter.New(Input=input_reader.GetOutput()) |
| 111 | +spcn_filter.SetColorIndexSuppressedByHematoxylin(0) |
| 112 | +spcn_filter.SetColorIndexSuppressedByEosin(1) |
| 113 | +spcn_filter.SetInput(0, input_reader.GetOutput()) |
| 114 | +spcn_filter.SetInput(1, reference_reader.GetOutput()) |
| 115 | + |
| 116 | +output_writer = itk.ImageFileWriter.New(spcn_filter.GetOutput()) |
| 117 | +output_writer.SetInput(spcn_filter.GetOutput()) |
| 118 | +output_writer.SetFileName(output_image_filename) |
| 119 | +output_writer.Write() |
| 120 | +``` |
| 121 | + |
| 122 | +Note that if `spcn_filter` is used again with a different `input_image`, for example from a different reader, |
| 123 | + |
| 124 | +```python |
| 125 | +spcn_filter.SetInput(0, input_reader2.GetOutput()) |
| 126 | +``` |
| 127 | + |
| 128 | +but the `reference_image` is unchanged then the filter will use its cached analysis of the `reference_image`, which |
| 129 | +saves about half the processing time. |
0 commit comments