diff --git a/.gitignore b/.gitignore
index b05281e..158faf3 100644
--- a/.gitignore
+++ b/.gitignore
@@ -127,4 +127,6 @@ docs/images/UML_diagrams/
docs/images/Graphical_abstract/
docs/images/Nfcore_module_figure
docs/presentations/
+docs/example_data/Earth_microbiome_vuegen_demo_notebook_test/
+docs/vuegen_case_study_earth_microbiome test.ipynb
test.py
\ No newline at end of file
diff --git a/README.md b/README.md
index 808ce35..c0aeac2 100644
--- a/README.md
+++ b/README.md
@@ -1,7 +1,7 @@
-
+
-----------------
VueGen is a Python library that automates the creation of scientific reports.
@@ -26,7 +26,7 @@ An overview of the VueGen workflow is shown in the figure below:

-->
-
+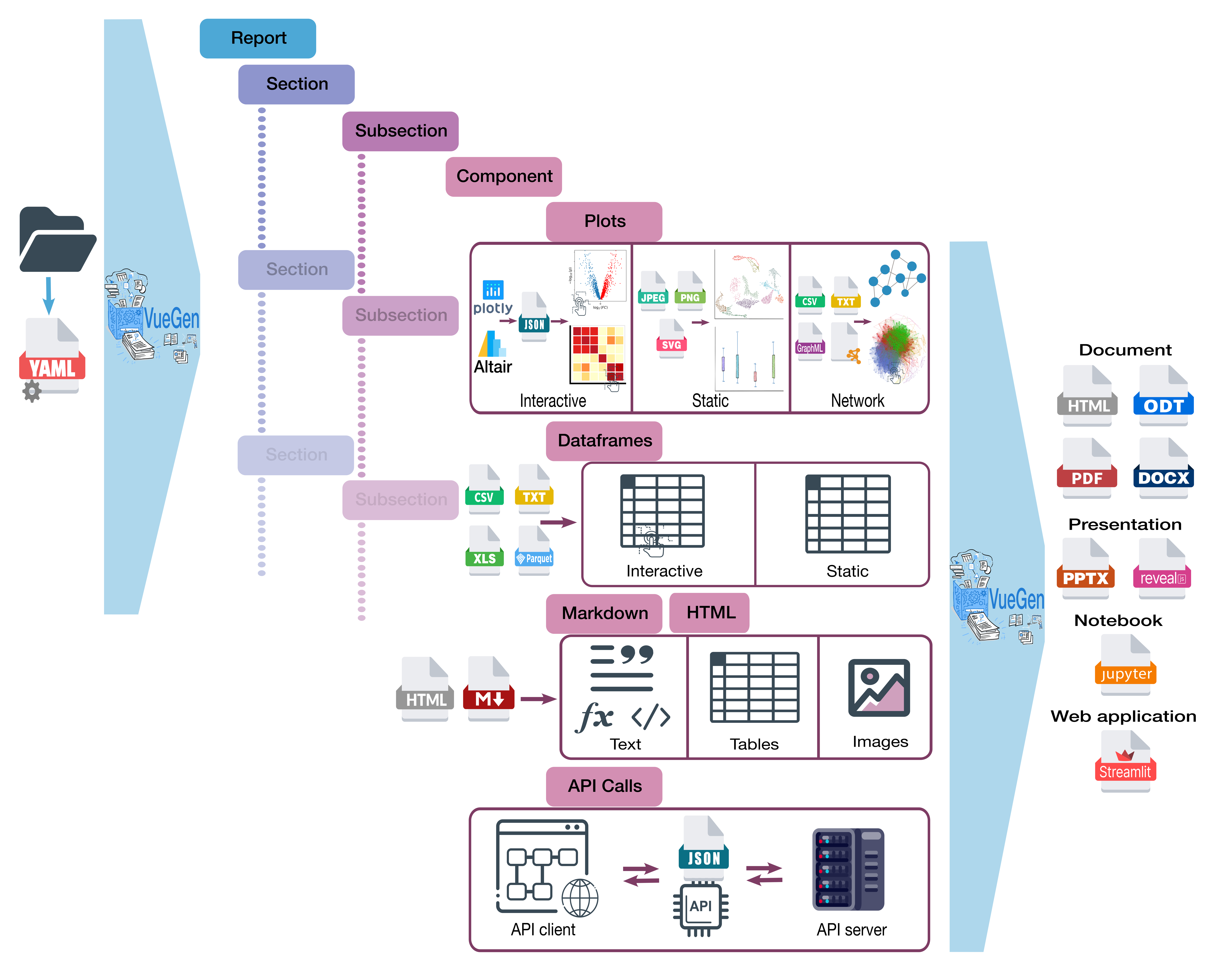
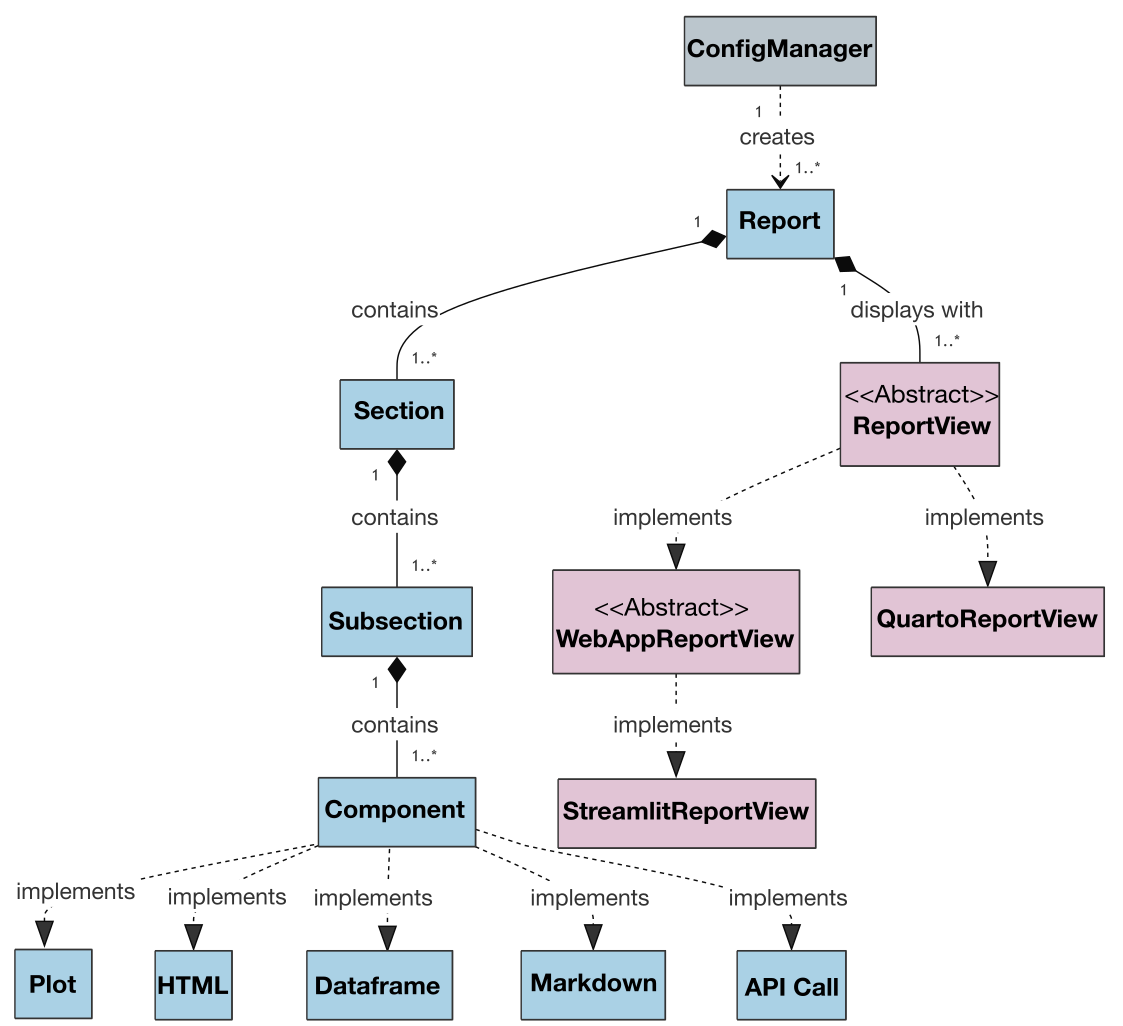
Also, the class diagram for the project is presented below to illustrate the architecture and relationships between classes:
@@ -36,33 +36,32 @@ Also, the class diagram for the project is presented below to illustrate the arc
-->
-
+
## Installation
-You can install the package for development from this repository by running the following command:
+Vuegen is available on [PyPI][vuegen-pypi] and can be installed using pip:
+
+```bash
+pip install vuegen
+```
+
+You can also install the package for development from this repository by running the following command:
```bash
pip install -e path/to/vuegen # specify location
pip install -e . # in case your pwd is in the vuegen directory
```
-This will both install `quarto` and `streamlit` as our backends for report generation.
-
-### Verify quarto installation
+### Quarto installation
-Test your quarto installation by running the following command:
+Vuegen uses [Quarto][quarto] to generate various report types. The pip insallation includes quarto using the [quarto-cli Python library][quarto-cli-pypi]. To test if quarto is installed in your computer, run the following command:
```bash
quarto check
```
-If you use conda a conda environement you can install quatro from the conda-forge channel
-in case it did not work.
-
-```bash
-conda install -c conda-forge quarto
-```
+If quarto is not installed, you can download the command-line interface from the [Quarto website][quarto-cli] for your operating system.
## Execution
@@ -83,7 +82,15 @@ It's also possible to provide a configuration file instead of a directory:
vuegen --config example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbiome_vuegen_demo_notebook_config.yaml --report_type streamlit
```
-The current report types are streamlit, html, pdf, docx, odt, revealjs, pptx, and jupyter.
+The current report types supported by VueGen are:
+* Streamlit
+* HTML
+* PDF
+* DOCX
+* ODT
+* Reveal.js
+* PPTX
+* Jupyter
## Acknowledgements
@@ -95,10 +102,15 @@ If you have comments or suggestions about this project, you can [open an issue][
[issues]: https://github.com/Multiomics-Analytics-Group/vuegen/issues/new
[streamlit]: https://streamlit.io/
+[vuegen-pypi]: https://pypi.org/project/vuegen/
+[quarto]: https://quarto.org/
+[quarto-cli-pypi]: https://pypi.org/project/quarto-cli/
+[quarto-cli]: https://quarto.org/docs/get-started/
[Mona]: https://multiomics-analytics-group.github.io/
[Biosustain]: https://www.biosustain.dtu.dk/
[scriberia]: https://www.scriberia.co.uk/
[turingway]: https://github.com/the-turing-way/the-turing-way
[zenodo-turingway]: https://zenodo.org/records/3695300
+[issues]: https://github.com/Multiomics-Analytics-Group/vuegen/issues/new
diff --git a/docs/images/vuegen_classdiagram_noattmeth.png b/docs/images/vuegen_classdiagram_noattmeth.png
index e9eeaa0..b7c578d 100644
Binary files a/docs/images/vuegen_classdiagram_noattmeth.png and b/docs/images/vuegen_classdiagram_noattmeth.png differ
diff --git a/docs/images/vuegen_graph_abstract.png b/docs/images/vuegen_graph_abstract.png
index caa114f..7d7fe84 100644
Binary files a/docs/images/vuegen_graph_abstract.png and b/docs/images/vuegen_graph_abstract.png differ
diff --git a/docs/images/vuegen_logo.png b/docs/images/vuegen_logo.png
index c753e09..5fdc22b 100644
Binary files a/docs/images/vuegen_logo.png and b/docs/images/vuegen_logo.png differ
diff --git a/docs/images/vuegen_logo.svg b/docs/images/vuegen_logo.svg
index a1df622..fff2065 100644
--- a/docs/images/vuegen_logo.svg
+++ b/docs/images/vuegen_logo.svg
@@ -11,8 +11,8 @@
sodipodi:docname="vuegen_logo.svg"
inkscape:version="1.3.2 (091e20e, 2023-11-25)"
inkscape:export-filename="vuegen_logo.png"
- inkscape:export-xdpi="100"
- inkscape:export-ydpi="100"
+ inkscape:export-xdpi="192"
+ inkscape:export-ydpi="192"
width="350"
height="300"
xmlns:inkscape="http://www.inkscape.org/namespaces/inkscape"
@@ -28,15 +28,15 @@
inkscape:pageopacity="0.0"
inkscape:pagecheckerboard="0"
inkscape:deskcolor="#d1d1d1"
- inkscape:zoom="0.89698939"
- inkscape:cx="146.04409"
- inkscape:cy="124.86212"
+ inkscape:zoom="1.2685346"
+ inkscape:cx="152.14406"
+ inkscape:cy="195.89533"
inkscape:window-width="1200"
- inkscape:window-height="847"
- inkscape:window-x="1201"
- inkscape:window-y="52"
+ inkscape:window-height="932"
+ inkscape:window-x="1200"
+ inkscape:window-y="25"
inkscape:window-maximized="0"
- inkscape:current-layer="g410"
+ inkscape:current-layer="Layer_1"
showgrid="true"
inkscape:document-units="mm">VueGen
-
+

