How to identify and remove dead end pores and isolated clusters after using reduce_coordination() #2567
-
Beta Was this translation helpful? Give feedback.
Replies: 3 comments
-
|
Isolate clusters is easy...these should show up on the |
Beta Was this translation helpful? Give feedback.
-
|
Oh, and I should also mention that trim coordination does not remove any pores, only throats, AND it does it in a way that maintains connectivity. It finds the "minimum spanning tree" of the network (assuming random throat weights), then only deletes throats that are NOT part of the spanning tree. |
Beta Was this translation helpful? Give feedback.
-
|
Thanks a lot, sir. It worked. |
Beta Was this translation helpful? Give feedback.
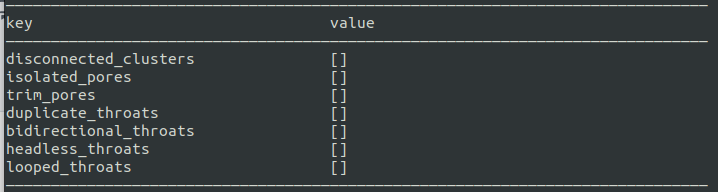
Isolate clusters is easy...these should show up on the
check_network_healthfunction. Deadend pores are not technically a problem for the simulations, so we consider those as healthy. However, if you really don't want deadend pores you can doPs = pn.num_neighbors(pores=np.Ps)which will return a list of the number of neighbors to each pore. You can then trim any pores with only 1 neighbor (op.topotools.trim(pn, pores=Ps==1). You'll need to do this several times since you may create new deadend pores upon deletion of the previous ones (as would happen in the case of a chain of pores leading to a dead end).