|
| 1 | +# Extended Physics-Informed Neural Networks (XPINNs) |
| 2 | + |
| 3 | +=== "模型训练命令" |
| 4 | + |
| 5 | + ``` sh |
| 6 | + # linux |
| 7 | + wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/XPINN/XPINN_2D_PoissonEqn.mat -P ./data/ |
| 8 | + |
| 9 | + # windows |
| 10 | + # curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/XPINN/XPINN_2D_PoissonEqn.mat --output ./data/XPINN_2D_PoissonEqn.mat |
| 11 | + |
| 12 | + python xpinn.py |
| 13 | + |
| 14 | + ``` |
| 15 | + |
| 16 | +## 1. 背景简介 |
| 17 | + |
| 18 | +求解偏微分方程(PDE)是一类基础的物理问题,随着人工智能技术的高速发展,利用深度学习求解偏微分方程成为新的研究趋势。[XPINNs(Extended Physics-Informed Neural Networks)](https://doi.org/10.4208/cicp.OA-2020-0164)是一种适用于物理信息神经网络(PINNs)的广义时空域分解方法,以求解任意复杂几何域上的非线性偏微分方程。 |
| 19 | + |
| 20 | +XPINNs 通过广义时空区域分解,有效地提高了模型的并行能力,并且支持高度不规则的、凸/非凸的时空域分解,界面条件是简单的。XPINNs 可扩展到任意类型的偏微分方程,而不论方程是何种物理性质。 |
| 21 | + |
| 22 | +精确求解高维复杂的方程已经成为科学计算的最大挑战之一,XPINNs 的优点使其成为模拟复杂方程的适用方法。 |
| 23 | + |
| 24 | +## 2. 问题定义 |
| 25 | + |
| 26 | +二维泊松方程: |
| 27 | + |
| 28 | +$$ \Delta u = f(x, y), x,y \in \Omega \subset R^2$$ |
| 29 | + |
| 30 | +## 3. 问题求解 |
| 31 | + |
| 32 | +接下来开始讲解如何将问题一步一步地转化为 PaddleScience 代码,用深度学习的方法求解该问题。 |
| 33 | +为了快速理解 PaddleScience,接下来仅对模型构建、方程构建、计算域构建等关键步骤进行阐述,而其余细节请参考 [API文档](../api/arch.md)。 |
| 34 | + |
| 35 | +### 3.1 数据集下载 |
| 36 | + |
| 37 | +如下图所示,数据集包含计算域的三个子区域的数据:红色区域的边界和残差点;黄色区域的界面;以及绿色区域的界面。 |
| 38 | + |
| 39 | +<figure markdown> |
| 40 | + 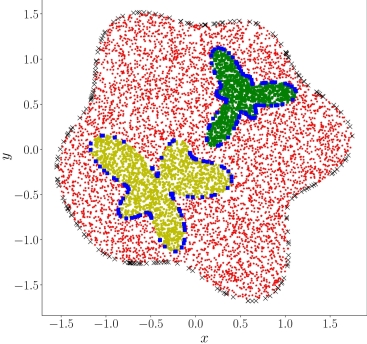 |
| 41 | + <figcaption>二维泊松方程的三个子区域</figcaption> |
| 42 | +</figure> |
| 43 | + |
| 44 | +计算域的边界表达式如下。 |
| 45 | + |
| 46 | +$$ \gamma =1.5+0.14 sin(4θ)+0.12 cos(6θ)+0.09 cos(5θ), θ \in [0,2π) $$ |
| 47 | + |
| 48 | +红色区域和黄色区域的界面的表达式如下。 |
| 49 | + |
| 50 | +$$ \gamma_1 =0.5+0.18 sin(3θ)+0.08 cos(2θ)+0.2 cos(5θ), θ \in [0,2π)$$ |
| 51 | + |
| 52 | +$$ \gamma_2 =0.34+0.04 sin(5θ)+0.18 cos(3θ)+0.1 cos(6θ), θ \in [0,2π) $$ |
| 53 | + |
| 54 | +执行以下命令,下载并解压数据集。 |
| 55 | + |
| 56 | +``` sh |
| 57 | +wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/XPINN/XPINN_2D_PoissonEqn.mat -P ./data/ |
| 58 | +``` |
| 59 | + |
| 60 | +### 3.3 模型构建 |
| 61 | + |
| 62 | +在本问题中,我们使用神经网络 `MLP` 作为模型,在模型代码中定义三个 `MLP` ,分别作为三个子区域的模型。 |
| 63 | + |
| 64 | +``` py linenums="301" |
| 65 | +--8<-- |
| 66 | +examples/xpinn/xpinn.py:301:302 |
| 67 | +--8<-- |
| 68 | +``` |
| 69 | + |
| 70 | +模型训练时,我们将使用 XPINN 方法分别计算每个子区域的模型损失。 |
| 71 | + |
| 72 | +<figure markdown> |
| 73 | + 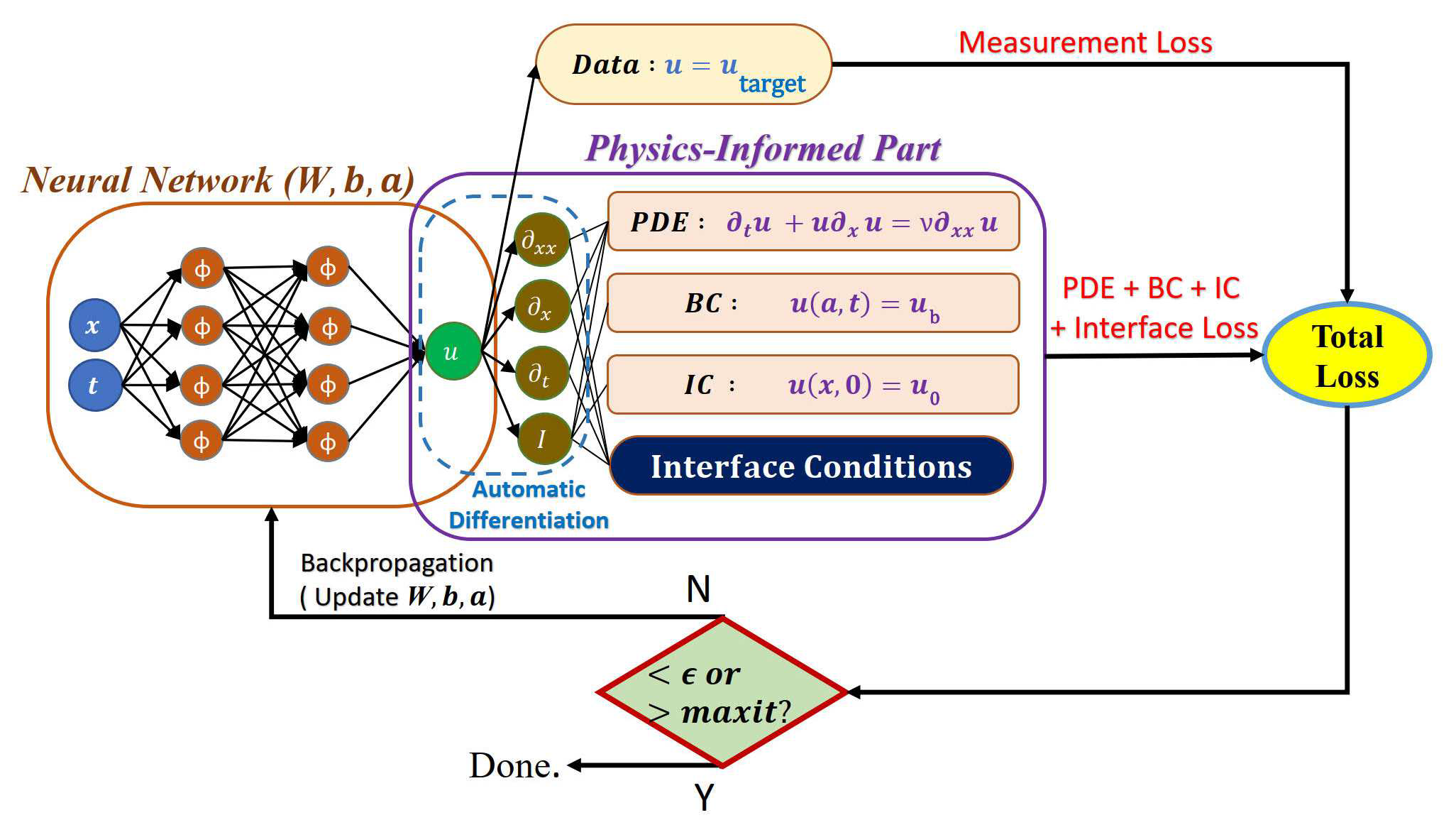 |
| 74 | + <figcaption>XPINN子网络的训练过程</figcaption> |
| 75 | +</figure> |
| 76 | + |
| 77 | +### 3.4 约束构建 |
| 78 | + |
| 79 | +在本案例中,我们使用监督数据集对模型进行训练,因此需要构建监督约束。 |
| 80 | + |
| 81 | +在定义约束之前,我们需要指定数据集的路径等相关配置,将这些信息存放到对应的 YAML 文件中,如下所示。 |
| 82 | + |
| 83 | +``` yaml linenums="43" |
| 84 | +--8<-- |
| 85 | +examples/xpinn/conf/xpinn.yaml:43:44 |
| 86 | +--8<-- |
| 87 | +``` |
| 88 | + |
| 89 | +接着定义训练损失函数的计算过程,调用 XPINN 方法计算损失,如下所示。 |
| 90 | + |
| 91 | +``` py linenums="130" |
| 92 | +--8<-- |
| 93 | +examples/xpinn/xpinn.py:130:191 |
| 94 | +--8<-- |
| 95 | +``` |
| 96 | + |
| 97 | +最后构建监督约束,如下所示。 |
| 98 | + |
| 99 | +``` py linenums="304" |
| 100 | +--8<-- |
| 101 | +examples/xpinn/xpinn.py:304:311 |
| 102 | +--8<-- |
| 103 | +``` |
| 104 | + |
| 105 | +### 3.5 超参数设定 |
| 106 | + |
| 107 | +设置训练轮数等参数,如下所示。 |
| 108 | + |
| 109 | +``` yaml linenums="83" |
| 110 | +--8<-- |
| 111 | +examples/xpinn/conf/xpinn.yaml:83:88 |
| 112 | +--8<-- |
| 113 | +``` |
| 114 | + |
| 115 | +### 3.6 优化器构建 |
| 116 | + |
| 117 | +训练过程会调用优化器来更新模型参数,此处选择较为常用的 `Adam` 优化器。 |
| 118 | + |
| 119 | +``` py linenums="337" |
| 120 | +--8<-- |
| 121 | +examples/xpinn/xpinn.py:337:338 |
| 122 | +--8<-- |
| 123 | +``` |
| 124 | + |
| 125 | +### 3.7 评估器构建 |
| 126 | + |
| 127 | +在训练过程中通常会按一定轮数间隔,用验证集(测试集)评估当前模型的训练情况,因此使用 `ppsci.validate.SupervisedValidator` 构建评估器。 |
| 128 | + |
| 129 | +``` py linenums="324" |
| 130 | +--8<-- |
| 131 | +examples/xpinn/xpinn.py:324:335 |
| 132 | +--8<-- |
| 133 | +``` |
| 134 | + |
| 135 | +评估指标为预测结果和真实结果的 RMSE 值,这里需自定义指标计算函数,如下所示。 |
| 136 | + |
| 137 | +``` py linenums="194" |
| 138 | +--8<-- |
| 139 | +examples/xpinn/xpinn.py:194:219 |
| 140 | +--8<-- |
| 141 | +``` |
| 142 | + |
| 143 | +### 3.8 模型训练 |
| 144 | + |
| 145 | +完成上述设置之后,只需要将上述实例化的对象按顺序传递给 `ppsci.solver.Solver`,然后启动训练。 |
| 146 | + |
| 147 | +``` py linenums="340" |
| 148 | +--8<-- |
| 149 | +examples/xpinn/xpinn.py:340:357 |
| 150 | +--8<-- |
| 151 | +``` |
| 152 | + |
| 153 | +### 3.9 结果可视化 |
| 154 | + |
| 155 | +训练完毕之后程序会对测试集中的数据进行预测,并以图片的形式对结果进行可视化,如下所示。 |
| 156 | + |
| 157 | +``` py linenums="360" |
| 158 | +--8<-- |
| 159 | +examples/xpinn/xpinn.py:360:384 |
| 160 | +--8<-- |
| 161 | +``` |
| 162 | + |
| 163 | +## 4. 完整代码 |
| 164 | + |
| 165 | +``` py linenums="1" title="cfdgcn.py" |
| 166 | +--8<-- |
| 167 | +examples/xpinn/xpinn.py |
| 168 | +--8<-- |
| 169 | +``` |
| 170 | + |
| 171 | +## 5. 结果展示 |
| 172 | + |
| 173 | +下方展示了对计算域中每个点的预测值结果、参考结果和相对误差。 |
| 174 | + |
| 175 | +<figure markdown> |
| 176 | + 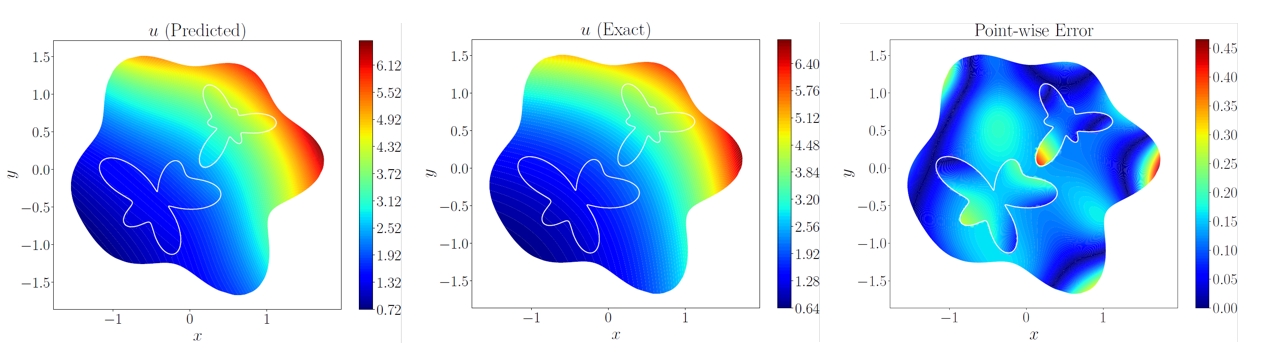 |
| 177 | + <figcaption>预测结果和参考结果的对比</figcaption> |
| 178 | +</figure> |
| 179 | + |
| 180 | +可以看到模型预测结果与真实结果相近,若增大训练轮数,模型精度会进一步提高。 |
| 181 | + |
| 182 | +## 6. 参考文献 |
| 183 | + |
| 184 | +* [A.D.Jagtap, G.E.Karniadakis, Extended Physics-Informed Neural Networks (XPINNs): A Generalized Space-Time Domain Decomposition Based Deep Learning Framework for Nonlinear Partial Differential Equations, Commun. Comput. Phys., Vol.28, No.5, 2002-2041, 2020.](https://doi.org/10.4208/cicp.OA-2020-0164) |
0 commit comments