diff --git a/README.md b/README.md
index 5b6ecba58d..4529c22442 100644
--- a/README.md
+++ b/README.md
@@ -124,6 +124,7 @@ PaddleScience 是一个基于深度学习框架 PaddlePaddle 开发的科学计
| 天气预报 | [FengWu 气象预报](https://paddlescience-docs.readthedocs.io/zh-cn/latest/zh/examples/fengwu) | 数据驱动 | Transformer | 监督学习 | - | [Paper](https://arxiv.org/pdf/2304.02948) |
| 天气预报 | [Pangu-Weather 气象预报](https://paddlescience-docs.readthedocs.io/zh-cn/latest/zh/examples/pangu_weather) | 数据驱动 | Transformer | 监督学习 | - | [Paper](https://arxiv.org/pdf/2211.02556) |
| 大气污染物 | [UNet 污染物扩散](https://aistudio.baidu.com/projectdetail/5663515?channel=0&channelType=0&sUid=438690&shared=1&ts=1698221963752) | 数据驱动 | UNet | 监督学习 | [Data](https://aistudio.baidu.com/datasetdetail/198102) | - |
+| 大气污染物 | [STAFNet 污染物浓度预测](https://paddlescience-docs.readthedocs.io/zh-cn/latest/zh/examples/stafnet) | 数据驱动 | STAFNet | 监督学习 | [Data](https://quotsoft.net/air) | [Paper](https://link.springer.com/chapter/10.1007/978-3-031-78186-5_22) |
| 天气预报 | [DGMR 气象预报](https://paddlescience-docs.readthedocs.io/zh-cn/latest/zh/examples/dgmr.md) | 数据驱动 | GAN | 监督学习 | [UK dataset](https://huggingface.co/datasets/openclimatefix/nimrod-uk-1km) | [Paper](https://arxiv.org/pdf/2104.00954.pdf) |
| 地震波形反演 | [VelocityGAN 地震波形反演](https://paddlescience-docs.readthedocs.io/zh-cn/latest/zh/examples/velocity_gan.md) | 数据驱动 | VelocityGAN | 监督学习 | [OpenFWI](https://openfwi-lanl.github.io/docs/data.html#vel) | [Paper](https://arxiv.org/abs/1809.10262v6) |
| 交通预测 | [TGCN 交通流量预测](https://paddlescience-docs.readthedocs.io/zh-cn/latest/zh/examples/tgcn.md) | 数据驱动 | GCN & CNN | 监督学习 | [PEMSD4 & PEMSD8](https://paddle-org.bj.bcebos.com/paddlescience/datasets/tgcn/tgcn_data.zip) | - |
diff --git a/docs/index.md b/docs/index.md
index 95ec7b42c5..5268a23fae 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -156,6 +156,7 @@
| 天气预报 | [FengWu 气象预报](./zh/examples/fengwu.md) | 数据驱动 | Transformer | 监督学习 | - | [Paper](https://arxiv.org/pdf/2304.02948) |
| 天气预报 | [Pangu-Weather 气象预报](./zh/examples/pangu_weather.md) | 数据驱动 | Transformer | 监督学习 | - | [Paper](https://arxiv.org/pdf/2211.02556) |
| 大气污染物 | [UNet 污染物扩散](https://aistudio.baidu.com/projectdetail/5663515?channel=0&channelType=0&sUid=438690&shared=1&ts=1698221963752) | 数据驱动 | UNet | 监督学习 | [Data](https://aistudio.baidu.com/datasetdetail/198102) | - |
+| 大气污染物 | [STAFNet 污染物浓度预测](./zh/examples/stafnet.md) | 数据驱动 | STAFNet | 监督学习 | [Data](https://quotsoft.net/air) | [Paper](https://link.springer.com/chapter/10.1007/978-3-031-78186-5_22) |
| 天气预报 | [DGMR 气象预报](./zh/examples/dgmr.md) | 数据驱动 | GAN | 监督学习 | [UK dataset](https://huggingface.co/datasets/openclimatefix/nimrod-uk-1km) | [Paper](https://arxiv.org/pdf/2104.00954.pdf) |
| 地震波形反演 | [VelocityGAN 地震波形反演](./zh/examples/velocity_gan.md) | 数据驱动 | VelocityGAN | 监督学习 | [OpenFWI](https://openfwi-lanl.github.io/docs/data.html#vel) | [Paper](https://arxiv.org/abs/1809.10262v6) |
| 交通预测 | [TGCN 交通流量预测](./zh/examples/tgcn.md) | 数据驱动 | GCN & CNN | 监督学习 | [PEMSD4 & PEMSD8](https://paddle-org.bj.bcebos.com/paddlescience/datasets/tgcn/tgcn_data.zip) | - |
diff --git a/docs/zh/api/arch.md b/docs/zh/api/arch.md
index e69bf9e752..456e7df97c 100644
--- a/docs/zh/api/arch.md
+++ b/docs/zh/api/arch.md
@@ -29,6 +29,7 @@
- NowcastNet
- SFNONet
- SPINN
+ - STAFNet
- TFNO1dNet
- TFNO2dNet
- TFNO3dNet
diff --git a/docs/zh/api/data/dataset.md b/docs/zh/api/data/dataset.md
index 60718dd0a3..e5a4c3d159 100644
--- a/docs/zh/api/data/dataset.md
+++ b/docs/zh/api/data/dataset.md
@@ -35,4 +35,5 @@
- DrivAerNetDataset
- DrivAerNetPlusPlusDataset
- IFMMoeDataset
+ - STAFNetDataset
show_root_heading: true
diff --git a/docs/zh/examples/stafnet.md b/docs/zh/examples/stafnet.md
new file mode 100644
index 0000000000..0ec8d7bf18
--- /dev/null
+++ b/docs/zh/examples/stafnet.md
@@ -0,0 +1,166 @@
+# STAFNet: Spatiotemporal-Aware Fusion Network for Air Quality Prediction
+
+| 预训练模型 | 指标 |
+| ------------------------------------------------------------ | -------------------- |
+| [stafnet.pdparams](https://paddle-org.bj.bcebos.com/paddlescience/models/stafnet/stafnet.pdparams) | MAE(1-48h) : 8.70933 |
+
+=== "模型训练命令"
+
+``` sh
+python stafnet.py DATASET.data_dir="Your train dataset path" EVAL.eval_data_path="Your evaluate dataset path"
+```
+
+=== "模型评估命令"
+
+``` sh
+wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/stafnet/val_data.pkl -P ./dataset/
+python stafnet.py mode=eval EVAL.pretrained_model_path="https://paddle-org.bj.bcebos.com/paddlescience/models/stafnet/stafnet.pdparams"
+```
+
+## 1. 背景介绍
+
+近些年,全球城市化和工业化不可避免地导致了严重的空气污染问题。心脏病、哮喘和肺癌等非传染性疾病的高发与暴露于空气污染直接相关。因此,空气质量预测已成为公共卫生、国民经济和城市管理的研究热点。目前已经建立了大量监测站来监测空气质量,并将其地理位置和历史观测数据合并为时空数据。然而,由于空气污染形成和扩散的高度复杂性,空气质量预测仍然面临着一些挑战。
+
+首先,空气中污染物的排放和扩散会导致邻近地区的空气质量迅速恶化,这一现象在托布勒地理第一定律中被描述为空间依赖关系,建立空间关系模型对于预测空气质量至关重要。然而,由于空气监测站的地理分布稀疏,要捕捉数据中内在的空间关联具有挑战性。其次,空气质量受到多源复杂因素的影响,尤其是气象条件。例如,长时间的小风或静风会抑制空气污染物的扩散,而自然降雨则在清除和冲刷空气污染物方面发挥作用。然而,空气质量站和气象站位于不同区域,导致多模态特征不对齐。融合不对齐的多模态特征并获取互补信息以准确预测空气质量是另一个挑战。最后但并非最不重要的一点是,空气质量的变化具有明显的多周期性特征。利用这一特点对提高空气质量预测的准确性非常重要,但也具有挑战性。
+
+针对空气质量预测提出了许多研究。早期的方法侧重于学习单个观测站观测数据的时间模式,而放弃了观测站之间的空间关系。最近,由于图神经网络(GNN)在处理非欧几里得图结构方面的有效性,越来越多的方法采用 GNN 来模拟空间依赖关系。这些方法将车站位置作为上下文特征,隐含地建立空间依赖关系模型,没有充分利用车站位置和车站之间关系所包含的宝贵空间信息。此外,现有的时空 GNN 缺乏在错位图中融合多个特征的能力。因此,大多数方法都需要额外的插值算法,以便在早期阶段将气象特征与 AQ 特征进行对齐和连接。这种方法消除了空气质量站和气象站之间的空间和结构信息,还可能引入噪声导致误差累积。此外,在空气质量预测中利用多周期性的问题仍未得到探索。
+
+该案例研究时空图网络在空气质量预测方向上的应用。
+
+## 2. 模型原理
+
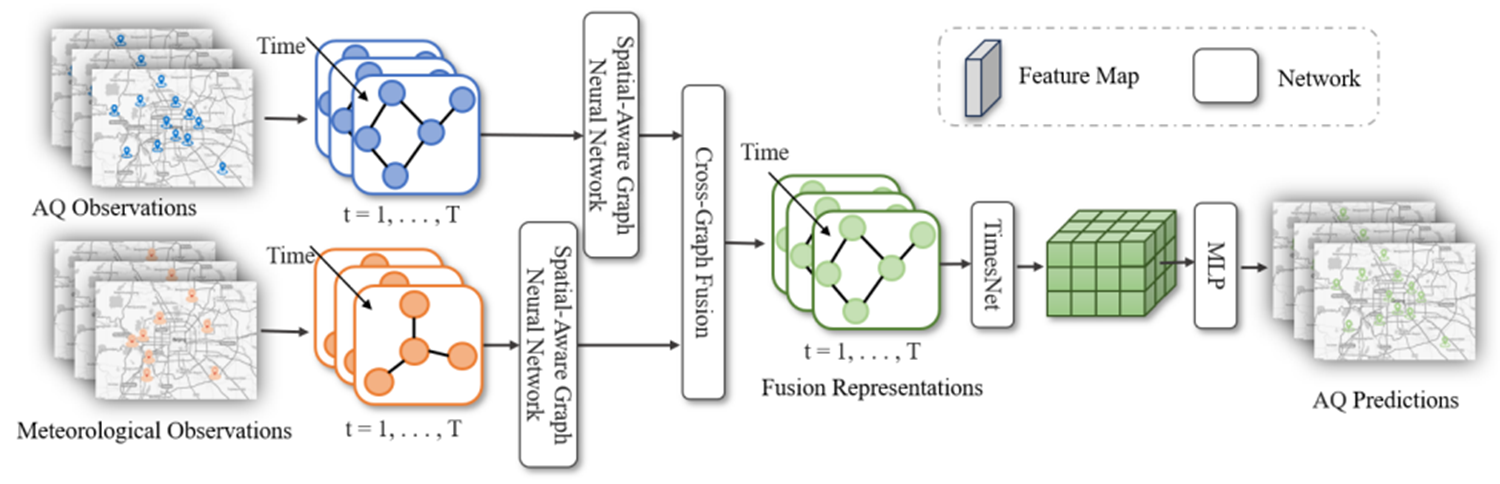
+STAFNet是一个新颖的多模式预报框架--时空感知融合网络来预测空气质量。STAFNet 由三个主要部分组成:空间感知 GNN、跨图融合关注机制和 TimesNet 。具体来说,为了捕捉各站点之间的空间关系,我们首先引入了空间感知 GNN,将空间信息明确纳入信息传递和节点表示中。为了全面表示气象影响,我们随后提出了一种基于交叉图融合关注机制的多模态融合策略,在不同类型站点的数量和位置不一致的情况下,将气象数据整合到 AQ 数据中。受多周期分析的启发,我们采用 TimesNet 将时间序列数据分解为不同频率的周期信号,并分别提取时间特征。
+
+本章节仅对 STAFNet的模型原理进行简单地介绍,详细的理论推导请阅读 STAFNet: Spatiotemporal-Aware Fusion Network for Air Quality Prediction
+
+模型的总体结构如图所示:
+
+
+
+STAFNet网络模型
+
+STAFNet 包含三个模块,分别将空间信息、气象信息和历史信息融合到空气质量特征表征中。首先模型的输入:过去T个时刻的**空气质量**数据和**气象**数据,使用两个空间感知 GNN(SAGNN),利用监测站之间的空间关系分别提取空气质量和气象信息。然后,跨图融合注意(CGF)将气象信息融合到空气质量表征中。最后,我们采用 TimesNet 模型来描述空气质量序列的时间动态,并生成多步骤预测。这一推理过程可表述如下,
+
+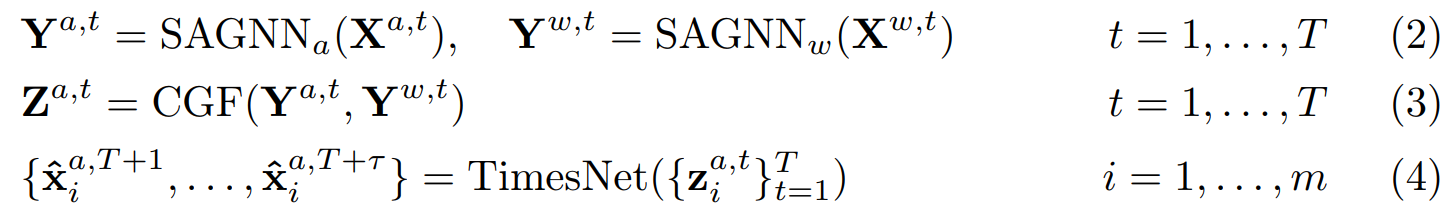
+
+## 3. 模型构建
+
+### 3.1 数据集介绍
+
+数据集采用了STAFNet处理好的北京空气质量数据集。数据集都包含:
+
+(1)空气质量观测值(即 PM2.5、PM10、O3、NO2、SO2 和 CO);
+
+(2)气象观测值(即温度、气压、湿度、风速和风向);
+
+(3)站点位置(即经度和纬度)。
+
+所有空气质量和气象观测数据每小时记录一次。数据集的收集时间为 2021 年 1 月 24 日至 2023 年 1 月 19 日,按 9:1的比例将数据分为训练集和测试集。空气质量观测数据来自国家城市空气质量实时发布平台,气象观测数据来自中国气象局。数据集的具体细节如下表所示:
+
+ 
北京空气质量数据集
+
+具体的数据集可从https://quotsoft.net/air/下载。
+
+运行本问题代码前请下载[数据集](https://paddle-org.bj.bcebos.com/paddlescience/datasets/stafnet/val_data.pkl), 下载后分别存放在路径:
+
+```
+./dataset
+```
+
+### 3.2 模型搭建
+
+在STAFNet模型中,输入过去72小时35个站点的空气质量数据,预测这35个站点未来48小时的空气质量。在本问题中,我们使用神经网络 `stafnet` 作为模型,其接收图结构数据,输出预测结果。
+
+```py linenums="10" title="examples/stafnet/stafnet.py"
+--8<--
+examples/stafnet/stafnet.py:10:10
+--8<--
+```
+
+### 3.4 参数和超参数设定
+
+其中超参数`cfg.MODEL.gat_hidden_dim`、`cfg.MODEL.e_layers`、`cfg.MODEL.d_model`、`cfg.MODEL.top_k`等默认设定如下:
+
+``` yaml linenums="35" title="examples/stafnet/conf/stafnet.yaml"
+--8<--
+examples/stafnet/conf/stafnet.yaml:35:59
+--8<--
+```
+
+### 3.5 优化器构建
+
+训练过程会调用优化器来更新模型参数,此处选择较为常用的 `Adam` 优化器。
+
+``` py linenums="62" title="examples/stafnet/stafnet.py"
+--8<--
+examples/stafnet/stafnet.py:62:62
+--8<--
+```
+
+其中学习率相关的设定如下:
+
+``` yaml linenums="70" title="examples/stafnet/conf/stafnet.yaml"
+--8<--
+examples/stafnet/conf/stafnet.yaml:70:75
+--8<--
+```
+
+### 3.6 约束构建
+
+在本案例中,我们使用监督数据集对模型进行训练,因此需要构建监督约束。
+
+在定义约束之前,我们需要指定数据集的路径等相关配置,将这些信息存放到对应的 YAML 文件中,如下所示。
+
+``` yaml linenums="31" title="examples/stafnet/conf/stafnet.yaml"
+--8<--
+examples/stafnet/conf/stafnet.yaml:31:34
+--8<--
+```
+
+最后构建监督约束,如下所示。
+
+``` py linenums="46" title="examples/stafnet/stafnet.py"
+--8<--
+examples/stafnet/stafnet.py:46:51
+--8<--
+```
+
+### 3.7 评估器构建
+
+在训练过程中通常会按一定轮数间隔,用验证集(测试集)评估当前模型的训练情况,因此使用 `ppsci.validate.SupervisedValidator` 构建评估器,构建过程与 [约束构建 3.6](https://github.com/PaddlePaddle/PaddleScience/blob/develop/docs/zh/examples/stafnet.md#36) 类似,只需把数据目录改为测试集的目录,并在配置文件中设置 `EVAL.batch_size=1` 即可。
+
+``` py linenums="52" title="examples/stafnet/stafnet.py"
+--8<--
+examples/stafnet/stafnet.py:52:58
+--8<--
+```
+
+评估指标为预测结果和真实结果的MAE 值,因此使用PaddleScience内置的`ppsci.metric.MAE()`,如下所示。
+
+``` py linenums="55" title="examples/stafnet/stafnet.py"
+--8<--
+examples/stafnet/stafnet.py:55:55
+--8<--
+```
+
+### 3.8 模型训练
+
+由于本问题为时序预测问题,因此可以使用PaddleScience内置的`psci.loss.MAELoss('mean')`作为训练过程的损失函数。同时选择使用随机梯度下降法对网络进行优化。完成述设置之后,只需要将上述实例化的对象按顺序传递给 `ppsci.solver.Solver`,然后启动训练。具体代码如下:
+
+``` py linenums="66" title="examples/stafnet/stafnet.py"
+--8<--
+examples/stafnet/stafnet.py:66:82
+--8<--
+```
+
+## 4. 完整代码
+
+```py linenums="1" title="examples/stafnet/stafnet.py"
+--8<--
+examples/stafnet/stafnet.py
+--8<--
+```
+
+## 5. 参考资料
+
+- [STAFNet: Spatiotemporal-Aware Fusion Network for Air Quality Prediction](https://link.springer.com/chapter/10.1007/978-3-031-78186-5_22)
diff --git a/examples/quick_start/case3.ipynb b/examples/quick_start/case3.ipynb
index 452502ceea..322e3832a9 100644
--- a/examples/quick_start/case3.ipynb
+++ b/examples/quick_start/case3.ipynb
@@ -81,7 +81,6 @@
"\n",
"import ppsci\n",
"import sympy as sp\n",
- "import numpy as np\n",
"\n",
"# 设置薄板计算域长、宽参数\n",
"Lx = 2.0 # 薄板x方向长度(m)\n",
diff --git a/examples/stafnet/conf/stafnet.yaml b/examples/stafnet/conf/stafnet.yaml
new file mode 100644
index 0000000000..87345cad66
--- /dev/null
+++ b/examples/stafnet/conf/stafnet.yaml
@@ -0,0 +1,84 @@
+defaults:
+ - ppsci_default
+ - TRAIN: train_default
+ - TRAIN/ema: ema_default
+ - TRAIN/swa: swa_default
+ - EVAL: eval_default
+ - INFER: infer_default
+ - hydra/job/config/override_dirname/exclude_keys: exclude_keys_default
+ - _self_
+hydra:
+ run:
+ # dynamic output directory according to running time and override name
+ dir: outputs_stafnet/${now:%Y-%m-%d}/${now:%H-%M-%S}/${hydra.job.override_dirname}
+ job:
+ name: ${mode} # name of logfile
+ chdir: false # keep current working directory unchanged
+ callbacks:
+ init_callback:
+ _target_: ppsci.utils.callbacks.InitCallback
+ sweep:
+ # output directory for multirun
+ dir: ${hydra.run.dir}
+ subdir: ./
+
+# general settings
+mode: train # running mode: train/eval
+seed: 42
+output_dir: ${hydra:run.dir}
+log_freq: 20
+# dataset setting
+DATASET:
+ label_keys: [label]
+ data_dir: ./dataset/train_data.pkl
+
+MODEL:
+ input_keys: [aq_train_data, mete_train_data]
+ output_keys: [label]
+ output_attention: true
+ seq_len: 72
+ pred_len: 48
+ aq_gat_node_features: 7
+ aq_gat_node_num: 35
+ mete_gat_node_features: 7
+ mete_gat_node_num: 18
+ gat_hidden_dim: 32
+ gat_edge_dim: 3
+ e_layers: 1
+ enc_in: 7
+ dec_in: 7
+ c_out: 7
+ d_model: 16
+ embed: fixed
+ freq: t
+ dropout: 0.05
+ factor: 3
+ n_heads: 4
+ d_ff: 32
+ num_kernels: 6
+ top_k: 4
+
+# training settings
+TRAIN:
+ epochs: 100
+ iters_per_epoch: 400
+ save_freq: 10
+ eval_during_train: true
+ eval_freq: 10
+ batch_size: 32
+ learning_rate: 0.0001
+ lr_scheduler:
+ epochs: ${TRAIN.epochs}
+ iters_per_epoch: ${TRAIN.iters_per_epoch}
+ learning_rate: 0.0005
+ step_size: 20
+ gamma: 0.95
+ pretrained_model_path: null
+ checkpoint_path: null
+
+EVAL:
+ eval_data_path: ./dataset/val_data.pkl
+ pretrained_model_path: null
+ compute_metric_by_batch: false
+ eval_with_no_grad: true
+ batch_size: 32
diff --git a/examples/stafnet/stafnet.py b/examples/stafnet/stafnet.py
new file mode 100644
index 0000000000..77f3c0160b
--- /dev/null
+++ b/examples/stafnet/stafnet.py
@@ -0,0 +1,139 @@
+import multiprocessing
+
+import hydra
+from omegaconf import DictConfig
+
+import ppsci
+
+
+def train(cfg: DictConfig):
+ # set model
+ model = ppsci.arch.STAFNet(**cfg.MODEL)
+ train_dataloader_cfg = {
+ "dataset": {
+ "name": "STAFNetDataset",
+ "file_path": cfg.DATASET.data_dir,
+ "input_keys": cfg.MODEL.input_keys,
+ "label_keys": cfg.MODEL.output_keys,
+ "seq_len": cfg.MODEL.seq_len,
+ "pred_len": cfg.MODEL.pred_len,
+ },
+ "batch_size": cfg.TRAIN.batch_size,
+ "sampler": {
+ "name": "BatchSampler",
+ "drop_last": False,
+ "shuffle": True,
+ },
+ "num_workers": 0,
+ }
+ eval_dataloader_cfg = {
+ "dataset": {
+ "name": "STAFNetDataset",
+ "file_path": cfg.EVAL.eval_data_path,
+ "input_keys": cfg.MODEL.input_keys,
+ "label_keys": cfg.MODEL.output_keys,
+ "seq_len": cfg.MODEL.seq_len,
+ "pred_len": cfg.MODEL.pred_len,
+ },
+ "batch_size": cfg.TRAIN.batch_size,
+ "sampler": {
+ "name": "BatchSampler",
+ "drop_last": False,
+ "shuffle": False,
+ },
+ "num_workers": 0,
+ }
+
+ sup_constraint = ppsci.constraint.SupervisedConstraint(
+ train_dataloader_cfg,
+ loss=ppsci.loss.MSELoss("mean"),
+ name="STAFNet_Sup",
+ )
+ constraint = {sup_constraint.name: sup_constraint}
+ sup_validator = ppsci.validate.SupervisedValidator(
+ eval_dataloader_cfg,
+ loss=ppsci.loss.MSELoss("mean"),
+ metric={"MAE": ppsci.metric.MAE()},
+ name="Sup_Validator",
+ )
+ validator = {sup_validator.name: sup_validator}
+
+ # set optimizer
+ lr_scheduler = ppsci.optimizer.lr_scheduler.Step(**cfg.TRAIN.lr_scheduler)()
+ optimizer = ppsci.optimizer.Adam(lr_scheduler)(model)
+ ITERS_PER_EPOCH = len(sup_constraint.data_loader)
+
+ # initialize solver
+ solver = ppsci.solver.Solver(
+ model,
+ constraint,
+ cfg.output_dir,
+ optimizer,
+ lr_scheduler,
+ cfg.TRAIN.epochs,
+ ITERS_PER_EPOCH,
+ eval_during_train=cfg.TRAIN.eval_during_train,
+ seed=cfg.seed,
+ validator=validator,
+ compute_metric_by_batch=cfg.EVAL.compute_metric_by_batch,
+ eval_with_no_grad=cfg.EVAL.eval_with_no_grad,
+ )
+
+ # train model
+ solver.train()
+
+
+def evaluate(cfg: DictConfig):
+ model = ppsci.arch.STAFNet(**cfg.MODEL)
+ eval_dataloader_cfg = {
+ "dataset": {
+ "name": "STAFNetDataset",
+ "file_path": cfg.EVAL.eval_data_path,
+ "input_keys": cfg.MODEL.input_keys,
+ "label_keys": cfg.MODEL.output_keys,
+ "seq_len": cfg.MODEL.seq_len,
+ "pred_len": cfg.MODEL.pred_len,
+ },
+ "batch_size": cfg.TRAIN.batch_size,
+ "sampler": {
+ "name": "BatchSampler",
+ "drop_last": False,
+ "shuffle": False,
+ },
+ "num_workers": 0,
+ }
+ sup_validator = ppsci.validate.SupervisedValidator(
+ eval_dataloader_cfg,
+ loss=ppsci.loss.MSELoss("mean"),
+ metric={"MAE": ppsci.metric.MAE()},
+ name="Sup_Validator",
+ )
+ validator = {sup_validator.name: sup_validator}
+
+ # initialize solver
+ solver = ppsci.solver.Solver(
+ model,
+ validator=validator,
+ cfg=cfg,
+ pretrained_model_path=cfg.EVAL.pretrained_model_path,
+ compute_metric_by_batch=cfg.EVAL.compute_metric_by_batch,
+ eval_with_no_grad=cfg.EVAL.eval_with_no_grad,
+ )
+
+ # evaluate model
+ solver.eval()
+
+
+@hydra.main(version_base=None, config_path="./conf", config_name="stafnet.yaml")
+def main(cfg: DictConfig):
+ if cfg.mode == "train":
+ train(cfg)
+ elif cfg.mode == "eval":
+ evaluate(cfg)
+ else:
+ raise ValueError(f"cfg.mode should in ['train', 'eval'], but got '{cfg.mode}'")
+
+
+if __name__ == "__main__":
+ multiprocessing.set_start_method("spawn")
+ main()
diff --git a/jointContribution/DU_CNN/requirements.txt b/jointContribution/DU_CNN/requirements.txt
index df2b9a3181..5a07c6511b 100644
--- a/jointContribution/DU_CNN/requirements.txt
+++ b/jointContribution/DU_CNN/requirements.txt
@@ -1,7 +1,7 @@
+mat73
+matplotlib
numpy
+paddlepaddle-gpu
pyaml
-tqdm
-matplotlib
sklearn
-mat73
-paddlepaddle-gpu
+tqdm
diff --git a/jointContribution/IJCAI_2024/aminos/download_dataset.ipynb b/jointContribution/IJCAI_2024/aminos/download_dataset.ipynb
index 48aa6357f8..8e60d995ff 100644
--- a/jointContribution/IJCAI_2024/aminos/download_dataset.ipynb
+++ b/jointContribution/IJCAI_2024/aminos/download_dataset.ipynb
@@ -1,139 +1,138 @@
{
- "cells": [
- {
- "cell_type": "markdown",
- "metadata": {
- "id": "kBRw5QHhBkax"
- },
- "source": [
- "# 数据集导入(预制链接)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {},
- "outputs": [],
- "source": [
- "\n",
- "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1JwR0Q1ArTg6c47EF2ZuIBpQwCPgXKrO2&export=download&authuser=0&confirm=t&uuid=8ce890e0-0019-4e1e-ac63-14718948f612&at=APZUnTW-e7sn7C7k5UVU2BaxZPGT%3A1721020888524' -O dataset_1.zip\n",
- "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1izP72pHtoXpQvOV8WFCnh_LekzLunyG5&export=download&authuser=0&confirm=t&uuid=8e453e3d-84ac-4f51-9cbf-45d47cbdcc65&at=APZUnTVfJYZBQwnHawB72aq5MPvv%3A1721020973099' -O dataset_2.zip\n",
- "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1djT0tlmLBi15LYZG0dxci1RSjPI94sM8&export=download&authuser=0&confirm=t&uuid=4687dd5d-a001-47f2-bacd-e72d5c7361e4&at=APZUnTWWEM2OCtpaZNuS4UQjMzxc%3A1721021154071' -O dataset_3.zip\n"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {
- "id": "gaD7ugivEL2R"
- },
- "source": [
- "## 官方版本数据导入"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {},
- "outputs": [],
- "source": [
- "# Unzip dataset file\n",
- "import os\n",
- "import shutil\n",
- "\n",
- "# 指定新文件夹的名称\n",
- "new_folder_A = 'unziped/dataset_1'\n",
- "new_folder_B = 'unziped/dataset_2'\n",
- "new_folder_C = 'unziped/dataset_3'\n",
- "\n",
- "# 在当前目录下创建新文件夹\n",
- "if not os.path.exists(new_folder_A):\n",
- " os.makedirs(new_folder_A)\n",
- "if not os.path.exists(new_folder_B):\n",
- " os.makedirs(new_folder_B)\n",
- "if not os.path.exists(new_folder_B):\n",
- " os.makedirs(new_folder_B)\n",
- "\n",
- "# 使用!unzip命令将文件解压到新文件夹中\n",
- "!unzip dataset_1.zip -d {new_folder_A}\n",
- "!unzip dataset_2.zip -d {new_folder_B}\n",
- "!unzip dataset_3.zip -d {new_folder_C}\n"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {
- "id": "z0Sek0wtEs5n"
- },
- "source": [
- "## 百度Baseline版本数据导入"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {
- "id": "kY81z-fCgPfK"
- },
- "source": [
- "## 自定义导入(在下面代码块导入并解压您的数据集)"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {
- "id": "ITzT8s2wgZG0"
- },
- "source": [
- "下载我们自己的数据集"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {},
- "outputs": [],
- "source": [
- "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1lUV7mJaoWPfRks4DMhJgDCxTZwMr8LQI&export=download&authuser=0&confirm=t&uuid=fbd3c896-23e8-42e0-b09d-410cf3c91487&at=APZUnTXNoF-geyh6R_qbWipUdgeP%3A1721020120231' -O trackCtrain.zip\n",
- "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1imPyFjFqAj5WTT_K-lHe7_6lFJ4azJ1L&export=download&authuser=0&confirm=t&uuid=ca9d62d3-394a-4798-b2f3-03455a381cf0&at=APZUnTVrYJw4okYQJSrdDXLPqdSX%3A1721020169107' -O trackCtest.zip\n",
- "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1y23xCHepK-NamTm3OFCAy41RnYolCjzq&export=download&authuser=0' -O trackCsrc.zip"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {},
- "outputs": [],
- "source": [
- "!mkdir ./Datasets\n",
- "!unzip trackCtrain.zip -d ./Datasets\n",
- "!unzip trackCtest.zip -d ./Datasets\n",
- "!unzip trackCsrc.zip"
- ]
- }
- ],
- "metadata": {
- "accelerator": "GPU",
- "colab": {
- "gpuType": "T4",
- "machine_shape": "hm",
- "provenance": []
- },
- "kernelspec": {
- "display_name": "Python 3",
- "name": "python3"
- },
- "language_info": {
- "codemirror_mode": {
- "name": "ipython",
- "version": 3
- },
- "file_extension": ".py",
- "mimetype": "text/x-python",
- "name": "python",
- "nbconvert_exporter": "python",
- "pygments_lexer": "ipython3",
- "version": "3.10.14"
- }
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {
+ "id": "kBRw5QHhBkax"
+ },
+ "source": [
+ "# 数据集导入(预制链接)"
+ ]
},
- "nbformat": 4,
- "nbformat_minor": 0
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "\n",
+ "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1JwR0Q1ArTg6c47EF2ZuIBpQwCPgXKrO2&export=download&authuser=0&confirm=t&uuid=8ce890e0-0019-4e1e-ac63-14718948f612&at=APZUnTW-e7sn7C7k5UVU2BaxZPGT%3A1721020888524' -O dataset_1.zip\n",
+ "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1izP72pHtoXpQvOV8WFCnh_LekzLunyG5&export=download&authuser=0&confirm=t&uuid=8e453e3d-84ac-4f51-9cbf-45d47cbdcc65&at=APZUnTVfJYZBQwnHawB72aq5MPvv%3A1721020973099' -O dataset_2.zip\n",
+ "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1djT0tlmLBi15LYZG0dxci1RSjPI94sM8&export=download&authuser=0&confirm=t&uuid=4687dd5d-a001-47f2-bacd-e72d5c7361e4&at=APZUnTWWEM2OCtpaZNuS4UQjMzxc%3A1721021154071' -O dataset_3.zip\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {
+ "id": "gaD7ugivEL2R"
+ },
+ "source": [
+ "## 官方版本数据导入"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Unzip dataset file\n",
+ "import os\n",
+ "\n",
+ "# 指定新文件夹的名称\n",
+ "new_folder_A = 'unziped/dataset_1'\n",
+ "new_folder_B = 'unziped/dataset_2'\n",
+ "new_folder_C = 'unziped/dataset_3'\n",
+ "\n",
+ "# 在当前目录下创建新文件夹\n",
+ "if not os.path.exists(new_folder_A):\n",
+ " os.makedirs(new_folder_A)\n",
+ "if not os.path.exists(new_folder_B):\n",
+ " os.makedirs(new_folder_B)\n",
+ "if not os.path.exists(new_folder_B):\n",
+ " os.makedirs(new_folder_B)\n",
+ "\n",
+ "# 使用!unzip命令将文件解压到新文件夹中\n",
+ "!unzip dataset_1.zip -d {new_folder_A}\n",
+ "!unzip dataset_2.zip -d {new_folder_B}\n",
+ "!unzip dataset_3.zip -d {new_folder_C}\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {
+ "id": "z0Sek0wtEs5n"
+ },
+ "source": [
+ "## 百度Baseline版本数据导入"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {
+ "id": "kY81z-fCgPfK"
+ },
+ "source": [
+ "## 自定义导入(在下面代码块导入并解压您的数据集)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {
+ "id": "ITzT8s2wgZG0"
+ },
+ "source": [
+ "下载我们自己的数据集"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1lUV7mJaoWPfRks4DMhJgDCxTZwMr8LQI&export=download&authuser=0&confirm=t&uuid=fbd3c896-23e8-42e0-b09d-410cf3c91487&at=APZUnTXNoF-geyh6R_qbWipUdgeP%3A1721020120231' -O trackCtrain.zip\n",
+ "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1imPyFjFqAj5WTT_K-lHe7_6lFJ4azJ1L&export=download&authuser=0&confirm=t&uuid=ca9d62d3-394a-4798-b2f3-03455a381cf0&at=APZUnTVrYJw4okYQJSrdDXLPqdSX%3A1721020169107' -O trackCtest.zip\n",
+ "!wget --no-check-certificate 'https://drive.usercontent.google.com/download?id=1y23xCHepK-NamTm3OFCAy41RnYolCjzq&export=download&authuser=0' -O trackCsrc.zip"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "!mkdir ./Datasets\n",
+ "!unzip trackCtrain.zip -d ./Datasets\n",
+ "!unzip trackCtest.zip -d ./Datasets\n",
+ "!unzip trackCsrc.zip"
+ ]
+ }
+ ],
+ "metadata": {
+ "accelerator": "GPU",
+ "colab": {
+ "gpuType": "T4",
+ "machine_shape": "hm",
+ "provenance": []
+ },
+ "kernelspec": {
+ "display_name": "Python 3",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.10.14"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 0
}
diff --git a/jointContribution/PINO/PINO_paddle/configs/pretrain/Darcy-pretrain.yaml b/jointContribution/PINO/PINO_paddle/configs/pretrain/Darcy-pretrain.yaml
index 7ebe16071f..6c5ce1935e 100644
--- a/jointContribution/PINO/PINO_paddle/configs/pretrain/Darcy-pretrain.yaml
+++ b/jointContribution/PINO/PINO_paddle/configs/pretrain/Darcy-pretrain.yaml
@@ -30,5 +30,3 @@ log:
project: 'PINO-Darcy-pretrain'
group: 'gelu-pino'
entity: hzzheng-pino
-
-
diff --git a/jointContribution/PINO/PINO_paddle/configs/pretrain/burgers-pretrain.yaml b/jointContribution/PINO/PINO_paddle/configs/pretrain/burgers-pretrain.yaml
index 7870982f5f..1b7ae25d4c 100644
--- a/jointContribution/PINO/PINO_paddle/configs/pretrain/burgers-pretrain.yaml
+++ b/jointContribution/PINO/PINO_paddle/configs/pretrain/burgers-pretrain.yaml
@@ -33,4 +33,3 @@ log:
project: PINO-burgers-pretrain
group: gelu-eqn
entity: hzzheng-pino
-
diff --git a/jointContribution/PINO/PINO_paddle/configs/test/burgers.yaml b/jointContribution/PINO/PINO_paddle/configs/test/burgers.yaml
index 716c8847fe..3314ea9e28 100644
--- a/jointContribution/PINO/PINO_paddle/configs/test/burgers.yaml
+++ b/jointContribution/PINO/PINO_paddle/configs/test/burgers.yaml
@@ -23,5 +23,3 @@ test:
log:
project: 'PINO-burgers-test'
group: 'gelu-test'
-
-
diff --git a/jointContribution/PINO/PINO_paddle/configs/test/darcy.yaml b/jointContribution/PINO/PINO_paddle/configs/test/darcy.yaml
index 9822de6122..1aa19178fb 100644
--- a/jointContribution/PINO/PINO_paddle/configs/test/darcy.yaml
+++ b/jointContribution/PINO/PINO_paddle/configs/test/darcy.yaml
@@ -22,5 +22,3 @@ test:
log:
project: 'PINO-Darcy'
group: 'default'
-
-
diff --git a/jointContribution/PINO/PINO_paddle/download_data.py b/jointContribution/PINO/PINO_paddle/download_data.py
index 5651617381..32f41d9ad4 100644
--- a/jointContribution/PINO/PINO_paddle/download_data.py
+++ b/jointContribution/PINO/PINO_paddle/download_data.py
@@ -1,40 +1,44 @@
import os
from argparse import ArgumentParser
+
import requests
from tqdm import tqdm
_url_dict = {
- 'NS-T4000': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fft_Re500_T4000.npy',
- 'NS-Re500Part0': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fine_Re500_T128_part0.npy',
- 'NS-Re500Part1': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fine_Re500_T128_part1.npy',
- 'NS-Re500Part2': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fine_Re500_T128_part2.npy',
- 'NS-Re100Part0': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fine_Re100_T128_part0.npy',
- 'burgers': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/burgers_pino.mat',
- 'NS-Re500_T300_id0': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/NS-Re500_T300_id0.npy',
- 'NS-Re500_T300_id0-shuffle': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS-Re500_T300_id0-shuffle.npy',
- 'darcy-train': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/piececonst_r421_N1024_smooth1.mat',
- 'Re500-1_8s-800-pino-140k': 'https://hkzdata.s3.us-west-2.amazonaws.com/PINO/checkpoints/Re500-1_8s-800-PINO-140000.pt',
+ "NS-T4000": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fft_Re500_T4000.npy",
+ "NS-Re500Part0": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fine_Re500_T128_part0.npy",
+ "NS-Re500Part1": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fine_Re500_T128_part1.npy",
+ "NS-Re500Part2": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fine_Re500_T128_part2.npy",
+ "NS-Re100Part0": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS_fine_Re100_T128_part0.npy",
+ "burgers": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/burgers_pino.mat",
+ "NS-Re500_T300_id0": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/NS-Re500_T300_id0.npy",
+ "NS-Re500_T300_id0-shuffle": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/NS-Re500_T300_id0-shuffle.npy",
+ "darcy-train": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/data/piececonst_r421_N1024_smooth1.mat",
+ "Re500-1_8s-800-pino-140k": "https://hkzdata.s3.us-west-2.amazonaws.com/PINO/checkpoints/Re500-1_8s-800-PINO-140000.pt",
}
+
def download_file(url, file_path):
- print('Start downloading...')
+ print("Start downloading...")
with requests.get(url, stream=True) as r:
r.raise_for_status()
- with open(file_path, 'wb') as f:
+ with open(file_path, "wb") as f:
for chunk in tqdm(r.iter_content(chunk_size=256 * 1024 * 1024)):
f.write(chunk)
- print('Complete')
+ print("Complete")
+
def main(args):
url = _url_dict[args.name]
- file_name = url.split('/')[-1]
+ file_name = url.split("/")[-1]
os.makedirs(args.outdir, exist_ok=True)
file_path = os.path.join(args.outdir, file_name)
download_file(url, file_path)
-if __name__ == '__main__':
- parser = ArgumentParser(description='Parser for downloading assets')
- parser.add_argument('--name', type=str, default='NS-T4000')
- parser.add_argument('--outdir', type=str, default='../data')
+
+if __name__ == "__main__":
+ parser = ArgumentParser(description="Parser for downloading assets")
+ parser.add_argument("--name", type=str, default="NS-T4000")
+ parser.add_argument("--outdir", type=str, default="../data")
args = parser.parse_args()
- main(args)
\ No newline at end of file
+ main(args)
diff --git a/jointContribution/PINO/PINO_paddle/eval_operator.py b/jointContribution/PINO/PINO_paddle/eval_operator.py
index 40991f86c9..6996381302 100644
--- a/jointContribution/PINO/PINO_paddle/eval_operator.py
+++ b/jointContribution/PINO/PINO_paddle/eval_operator.py
@@ -1,78 +1,94 @@
-import yaml
+from argparse import ArgumentParser
import paddle
+import yaml
+from models import FNO2d
+from models import FNO3d
from paddle.io import DataLoader
-from models import FNO3d, FNO2d
-from train_utils import NSLoader, get_forcing, DarcyFlow
-
-from train_utils.eval_3d import eval_ns
+from train_utils import DarcyFlow
+from train_utils import NSLoader
+from train_utils import get_forcing
from train_utils.eval_2d import eval_darcy
+from train_utils.eval_3d import eval_ns
-from argparse import ArgumentParser
def test_3d(config):
- device = 0 if paddle.cuda.is_available() else 'cpu'
- data_config = config['data']
- loader = NSLoader(datapath1=data_config['datapath'],
- nx=data_config['nx'], nt=data_config['nt'],
- sub=data_config['sub'], sub_t=data_config['sub_t'],
- N=data_config['total_num'],
- t_interval=data_config['time_interval'])
+ device = 0 if paddle.cuda.is_available() else "cpu"
+ data_config = config["data"]
+ loader = NSLoader(
+ datapath1=data_config["datapath"],
+ nx=data_config["nx"],
+ nt=data_config["nt"],
+ sub=data_config["sub"],
+ sub_t=data_config["sub_t"],
+ N=data_config["total_num"],
+ t_interval=data_config["time_interval"],
+ )
- eval_loader = loader.make_loader(n_sample=data_config['n_sample'],
- batch_size=config['test']['batchsize'],
- start=data_config['offset'],
- train=data_config['shuffle'])
- model = FNO3d(modes1=config['model']['modes1'],
- modes2=config['model']['modes2'],
- modes3=config['model']['modes3'],
- fc_dim=config['model']['fc_dim'],
- layers=config['model']['layers']).to(device)
+ eval_loader = loader.make_loader(
+ n_sample=data_config["n_sample"],
+ batch_size=config["test"]["batchsize"],
+ start=data_config["offset"],
+ train=data_config["shuffle"],
+ )
+ model = FNO3d(
+ modes1=config["model"]["modes1"],
+ modes2=config["model"]["modes2"],
+ modes3=config["model"]["modes3"],
+ fc_dim=config["model"]["fc_dim"],
+ layers=config["model"]["layers"],
+ ).to(device)
- if 'ckpt' in config['test']:
- ckpt_path = config['test']['ckpt']
+ if "ckpt" in config["test"]:
+ ckpt_path = config["test"]["ckpt"]
ckpt = paddle.load(ckpt_path)
- model.load_state_dict(ckpt['model'])
- print('Weights loaded from %s' % ckpt_path)
- print(f'Resolution : {loader.S}x{loader.S}x{loader.T}')
+ model.load_state_dict(ckpt["model"])
+ print("Weights loaded from %s" % ckpt_path)
+ print(f"Resolution : {loader.S}x{loader.S}x{loader.T}")
forcing = get_forcing(loader.S).to(device)
- eval_ns(model,
- loader,
- eval_loader,
- forcing,
- config,
- device=device)
+ eval_ns(model, loader, eval_loader, forcing, config, device=device)
+
def test_2d(config):
- data_config = config['data']
- dataset = DarcyFlow(data_config['datapath'],
- nx=data_config['nx'], sub=data_config['sub'],
- offset=data_config['offset'], num=data_config['n_sample'])
- dataloader = DataLoader(dataset, batch_size=config['test']['batchsize'], shuffle=False)
- model = FNO2d(modes1=config['model']['modes1'],
- modes2=config['model']['modes2'],
- fc_dim=config['model']['fc_dim'],
- layers=config['model']['layers'],
- act=config['model']['act'])
+ data_config = config["data"]
+ dataset = DarcyFlow(
+ data_config["datapath"],
+ nx=data_config["nx"],
+ sub=data_config["sub"],
+ offset=data_config["offset"],
+ num=data_config["n_sample"],
+ )
+ dataloader = DataLoader(
+ dataset, batch_size=config["test"]["batchsize"], shuffle=False
+ )
+ model = FNO2d(
+ modes1=config["model"]["modes1"],
+ modes2=config["model"]["modes2"],
+ fc_dim=config["model"]["fc_dim"],
+ layers=config["model"]["layers"],
+ act=config["model"]["act"],
+ )
# Load from checkpoint
- if 'ckpt' in config['test']:
- ckpt_path = config['test']['ckpt']
+ if "ckpt" in config["test"]:
+ ckpt_path = config["test"]["ckpt"]
ckpt = paddle.load(ckpt_path)
- model.set_dict(ckpt['model'])
- print('Weights loaded from %s' % ckpt_path)
+ model.set_dict(ckpt["model"])
+ print("Weights loaded from %s" % ckpt_path)
eval_darcy(model, dataloader, config)
-if __name__ == '__main__':
- parser = ArgumentParser(description='Basic paser')
- parser.add_argument('--config_path', type=str, help='Path to the configuration file')
- parser.add_argument('--log', action='store_true', help='Turn on the wandb')
+
+if __name__ == "__main__":
+ parser = ArgumentParser(description="Basic paser")
+ parser.add_argument(
+ "--config_path", type=str, help="Path to the configuration file"
+ )
+ parser.add_argument("--log", action="store_true", help="Turn on the wandb")
options = parser.parse_args()
config_file = options.config_path
- with open(config_file, 'r') as stream:
+ with open(config_file, "r") as stream:
config = yaml.load(stream, yaml.FullLoader)
- if 'name' in config['data'] and config['data']['name'] == 'Darcy':
+ if "name" in config["data"] and config["data"]["name"] == "Darcy":
test_2d(config)
else:
test_3d(config)
-
diff --git a/jointContribution/PINO/PINO_paddle/models/FCN.py b/jointContribution/PINO/PINO_paddle/models/FCN.py
index d429d0f43f..7435c76eca 100644
--- a/jointContribution/PINO/PINO_paddle/models/FCN.py
+++ b/jointContribution/PINO/PINO_paddle/models/FCN.py
@@ -1,29 +1,31 @@
import paddle.nn as nn
+
def linear_block(in_channel, out_channel):
- block = nn.Sequential(
- nn.Linear(in_channel, out_channel),
- nn.Tanh()
- )
+ block = nn.Sequential(nn.Linear(in_channel, out_channel), nn.Tanh())
return block
+
class FCNet(nn.Layer):
- '''
+ """
Fully connected layers with Tanh as nonlinearity
Reproduced from PINNs Burger equation
- '''
+ """
def __init__(self, layers=[2, 10, 1]):
super(FCNet, self).__init__()
- fc_list = [linear_block(in_size, out_size)
- for in_size, out_size in zip(layers, layers[1:-1])]
+ fc_list = [
+ linear_block(in_size, out_size)
+ for in_size, out_size in zip(layers, layers[1:-1])
+ ]
fc_list.append(nn.Linear(layers[-2], layers[-1]))
self.fc = nn.Sequential(*fc_list)
def forward(self, x):
return self.fc(x)
+
class DenseNet(nn.Layer):
def __init__(self, layers, nonlinearity, out_nonlinearity=None, normalize=False):
super(DenseNet, self).__init__()
@@ -31,20 +33,20 @@ def __init__(self, layers, nonlinearity, out_nonlinearity=None, normalize=False)
self.n_layers = len(layers) - 1
assert self.n_layers >= 1
if isinstance(nonlinearity, str):
- if nonlinearity == 'tanh':
+ if nonlinearity == "tanh":
nonlinearity = nn.Tanh
- elif nonlinearity == 'relu':
+ elif nonlinearity == "relu":
nonlinearity == nn.ReLU
else:
- raise ValueError(f'{nonlinearity} is not supported')
+ raise ValueError(f"{nonlinearity} is not supported")
self.layers = nn.ModuleList()
for j in range(self.n_layers):
- self.layers.append(nn.Linear(layers[j], layers[j+1]))
+ self.layers.append(nn.Linear(layers[j], layers[j + 1]))
if j != self.n_layers - 1:
if normalize:
- self.layers.append(nn.BatchNorm1d(layers[j+1]))
+ self.layers.append(nn.BatchNorm1d(layers[j + 1]))
self.layers.append(nonlinearity())
@@ -56,4 +58,3 @@ def forward(self, x):
x = l(x)
return x
-
diff --git a/jointContribution/PINO/PINO_paddle/models/FNO_blocks.py b/jointContribution/PINO/PINO_paddle/models/FNO_blocks.py
index c158f4f217..86ff9a8241 100644
--- a/jointContribution/PINO/PINO_paddle/models/FNO_blocks.py
+++ b/jointContribution/PINO/PINO_paddle/models/FNO_blocks.py
@@ -1,9 +1,11 @@
+import itertools
+
import paddle
import paddle.nn as nn
-import itertools
einsum_symbols = "abcdefghijklmnopqrstuvwxyzABCDEFGHIJKLMNOPQRSTUVWXYZ"
+
def _contract_dense(x, weight, separable=False):
# order = tl.ndim(x)
order = len(x.shape)
@@ -28,6 +30,7 @@ def _contract_dense(x, weight, separable=False):
return paddle.einsum(eq, x, weight)
+
def _contract_dense_trick(x, weight, separable=False):
# the same as above function, but do the complex multiplication manually to avoid the einsum bug in paddle
weight_real = weight.data.real()
@@ -59,11 +62,13 @@ def _contract_dense_trick(x, weight, separable=False):
x = paddle.complex(o1_real, o1_imag)
return x
+
def _contract_dense_separable(x, weight, separable=True):
- if separable == False:
+ if not separable:
raise ValueError("This function is only for separable=True")
return x * weight
+
def _contract_cp(x, cp_weight, separable=False):
# order = tl.ndim(x)
order = len(x.shape)
@@ -84,6 +89,7 @@ def _contract_cp(x, cp_weight, separable=False):
return paddle.einsum(eq, x, cp_weight.weights, *cp_weight.factors)
+
def _contract_tucker(x, tucker_weight, separable=False):
# order = tl.ndim(x)
order = len(x.shape)
@@ -118,6 +124,7 @@ def _contract_tucker(x, tucker_weight, separable=False):
print(eq) # 'abcd,fghi,bf,eg,ch,di->aecd'
return paddle.einsum(eq, x, tucker_weight.core, *tucker_weight.factors)
+
def _contract_tt(x, tt_weight, separable=False):
# order = tl.ndim(x)
order = len(x.shape)
@@ -144,16 +151,17 @@ def _contract_tt(x, tt_weight, separable=False):
return paddle.einsum(eq, x, *tt_weight.factors)
+
def get_contract_fun(weight, implementation="reconstructed", separable=False):
"""Generic ND implementation of Fourier Spectral Conv contraction
-
+
Parameters
----------
weight : tensorl-paddle's FactorizedTensor
implementation : {'reconstructed', 'factorized'}, default is 'reconstructed'
whether to reconstruct the weight and do a forward pass (reconstructed)
or contract directly the factors of the factorized weight with the input (factorized)
-
+
Returns
-------
function : (x, weight) -> x * weight in Fourier space
@@ -176,6 +184,7 @@ def get_contract_fun(weight, implementation="reconstructed", separable=False):
f'Got implementation = {implementation}, expected "reconstructed" or "factorized"'
)
+
class FactorizedTensor(nn.Layer):
def __init__(self, shape, init_scale):
super().__init__()
@@ -195,6 +204,7 @@ def __repr__(self):
def data(self):
return paddle.complex(self.real, self.imag)
+
class FactorizedSpectralConv(nn.Layer):
"""Generic N-Dimensional Fourier Neural Operator
@@ -258,7 +268,7 @@ def __init__(
if isinstance(n_modes, int):
n_modes = [n_modes]
self.n_modes = n_modes
- half_modes = [m // 2 for m in n_modes]
+ [m // 2 for m in n_modes]
self.half_modes = n_modes
self.rank = rank
@@ -324,14 +334,14 @@ def __init__(
def forward(self, x, indices=0):
"""Generic forward pass for the Factorized Spectral Conv
-
- Parameters
+
+ Parameters
----------
x : paddle.Tensor
input activation of size (batch_size, channels, d1, ..., dN)
indices : int, default is 0
if joint_factorization, index of the layers for n_layers > 1
-
+
Returns
-------
tensorized_spectral_conv(x)
@@ -348,7 +358,8 @@ def forward(self, x, indices=0):
x = paddle.fft.rfftn(x_float, norm=self.fft_norm, axes=fft_dims)
out_fft = paddle.zeros(
- [batchsize, self.out_channels, *fft_size], dtype=paddle.complex64,
+ [batchsize, self.out_channels, *fft_size],
+ dtype=paddle.complex64,
) # [1,32,16,9], all zeros, complex
# We contract all corners of the Fourier coefs
@@ -402,13 +413,14 @@ def get_conv(self, indices):
def __getitem__(self, indices):
return self.get_conv(indices)
+
class SubConv2d(nn.Layer):
"""Class representing one of the convolutions from the mother joint factorized convolution
Notes
-----
This relies on the fact that nn.Parameters are not duplicated:
- if the same nn.Parameter is assigned to multiple modules, they all point to the same data,
+ if the same nn.Parameter is assigned to multiple modules, they all point to the same data,
which is shared.
"""
@@ -420,6 +432,7 @@ def __init__(self, main_conv, indices):
def forward(self, x):
return self.main_conv.forward(x, self.indices)
+
class FactorizedSpectralConv1d(FactorizedSpectralConv):
def __init__(
self,
@@ -462,7 +475,8 @@ def forward(self, x, indices=0):
x = paddle.fft.rfft(x, norm=self.fft_norm)
out_fft = paddle.zeros(
- [batchsize, self.out_channels, width // 2 + 1], dtype=paddle.complex64,
+ [batchsize, self.out_channels, width // 2 + 1],
+ dtype=paddle.complex64,
)
out_fft[:, :, : self.half_modes_height] = self._contract(
x[:, :, : self.half_modes_height],
@@ -477,6 +491,7 @@ def forward(self, x, indices=0):
return x
+
class FactorizedSpectralConv2d(FactorizedSpectralConv):
def __init__(
self,
@@ -522,7 +537,8 @@ def forward(self, x, indices=0):
# The output will be of size (batch_size, self.out_channels, x.size(-2), x.size(-1)//2 + 1)
out_fft = paddle.zeros(
- [batchsize, self.out_channels, height, width // 2 + 1], dtype=x.dtype,
+ [batchsize, self.out_channels, height, width // 2 + 1],
+ dtype=x.dtype,
)
# upper block (truncate high freq)
@@ -551,6 +567,7 @@ def forward(self, x, indices=0):
return x
+
class FactorizedSpectralConv3d(FactorizedSpectralConv):
def __init__(
self,
@@ -679,6 +696,7 @@ def forward(self, x, indices=0):
return x
+
if __name__ == "__main__":
# let x be a complex tensor of size (32, 32, 8, 8)
x = paddle.randn([32, 32, 8, 8]).astype("complex64")
diff --git a/jointContribution/PINO/PINO_paddle/models/basics.py b/jointContribution/PINO/PINO_paddle/models/basics.py
index 8a8fea46f3..73bfdd57b1 100644
--- a/jointContribution/PINO/PINO_paddle/models/basics.py
+++ b/jointContribution/PINO/PINO_paddle/models/basics.py
@@ -1,33 +1,36 @@
-import numpy as np
-
import paddle
import paddle.nn as nn
import paddle.nn.initializer as Initializer
+
def compl_mul1d(a: paddle.Tensor, b: paddle.Tensor) -> paddle.Tensor:
# (batch, in_channel, x ), (in_channel, out_channel, x) -> (batch, out_channel, x)
res = paddle.einsum("bix,iox->box", a, b)
return res
+
def compl_mul2d(a: paddle.Tensor, b: paddle.Tensor) -> paddle.Tensor:
# (batch, in_channel, x,y,t ), (in_channel, out_channel, x,y,t) -> (batch, out_channel, x,y,t)
- res = paddle.einsum("bixy,ioxy->boxy", a, b)
+ res = paddle.einsum("bixy,ioxy->boxy", a, b)
return res
+
def compl_mul3d(a: paddle.Tensor, b: paddle.Tensor) -> paddle.Tensor:
res = paddle.einsum("bixyz,ioxyz->boxyz", a, b)
return res
+
################################################################
# 1d fourier layer
################################################################
+
class SpectralConv1d(nn.Layer):
def __init__(self, in_channels, out_channels, modes1):
super(SpectralConv1d, self).__init__()
"""
- 1D Fourier layer. It does FFT, linear transform, and Inverse FFT.
+ 1D Fourier layer. It does FFT, linear transform, and Inverse FFT.
"""
self.in_channels = in_channels
@@ -35,8 +38,14 @@ def __init__(self, in_channels, out_channels, modes1):
# Number of Fourier modes to multiply, at most floor(N/2) + 1
self.modes1 = modes1
- self.scale = (1 / (in_channels*out_channels))
- self.weights1 = paddle.create_parameter(shape=(in_channels, out_channels), dtype=paddle.complex64, attr=Initializer.Assign(self.scale * paddle.rand(in_channels, out_channels, self.modes1)))
+ self.scale = 1 / (in_channels * out_channels)
+ self.weights1 = paddle.create_parameter(
+ shape=(in_channels, out_channels),
+ dtype=paddle.complex64,
+ attr=Initializer.Assign(
+ self.scale * paddle.rand(in_channels, out_channels, self.modes1)
+ ),
+ )
def forward(self, x):
batchsize = x.shape[0]
@@ -44,17 +53,23 @@ def forward(self, x):
x_ft = paddle.fft.rfftn(x, dim=[2])
# Multiply relevant Fourier modes
- out_ft = paddle.zeros(batchsize, self.in_channels, x.shape[-1]//2 + 1, dtype=paddle.complex64)
- out_ft[:, :, :self.modes1] = compl_mul1d(x_ft[:, :, :self.modes1], self.weights1)
+ out_ft = paddle.zeros(
+ batchsize, self.in_channels, x.shape[-1] // 2 + 1, dtype=paddle.complex64
+ )
+ out_ft[:, :, : self.modes1] = compl_mul1d(
+ x_ft[:, :, : self.modes1], self.weights1
+ )
# Return to physical space
x = paddle.fft.irfftn(out_ft, s=[x.shape[-1]], axes=[2])
return x
+
################################################################
# 2d fourier layer
################################################################
+
class SpectralConv2d(nn.Layer):
def __init__(self, in_channels, out_channels, modes1, modes2):
super(SpectralConv2d, self).__init__()
@@ -64,102 +79,212 @@ def __init__(self, in_channels, out_channels, modes1, modes2):
self.modes1 = modes1
self.modes2 = modes2
- self.scale = (1 / (in_channels * out_channels))
- weights1_attr = Initializer.Assign(self.scale * paddle.rand([in_channels, out_channels, self.modes1, self.modes2], dtype=paddle.float32))
- self.weights1_real = paddle.create_parameter(shape=(in_channels, out_channels), dtype=paddle.float32, attr=weights1_attr)
- self.weights1_imag = paddle.create_parameter(shape=(in_channels, out_channels), dtype=paddle.float32, attr=weights1_attr)
+ self.scale = 1 / (in_channels * out_channels)
+ weights1_attr = Initializer.Assign(
+ self.scale
+ * paddle.rand(
+ [in_channels, out_channels, self.modes1, self.modes2],
+ dtype=paddle.float32,
+ )
+ )
+ self.weights1_real = paddle.create_parameter(
+ shape=(in_channels, out_channels), dtype=paddle.float32, attr=weights1_attr
+ )
+ self.weights1_imag = paddle.create_parameter(
+ shape=(in_channels, out_channels), dtype=paddle.float32, attr=weights1_attr
+ )
self.weights1 = paddle.concat([self.weights1_real, self.weights1_imag], axis=0)
- self.weights2 = paddle.create_parameter(shape=(in_channels, out_channels), dtype=paddle.float32, attr=Initializer.Assign(self.scale * paddle.rand([in_channels, out_channels, self.modes1, self.modes2], dtype=paddle.float32)))
+ self.weights2 = paddle.create_parameter(
+ shape=(in_channels, out_channels),
+ dtype=paddle.float32,
+ attr=Initializer.Assign(
+ self.scale
+ * paddle.rand(
+ [in_channels, out_channels, self.modes1, self.modes2],
+ dtype=paddle.float32,
+ )
+ ),
+ )
def forward(self, x):
batchsize = x.shape[0]
- size1 = x.shape[-2]
- size2 = x.shape[-1]
+ x.shape[-2]
+ x.shape[-1]
# Compute Fourier coeffcients up to factor of e^(- something constant)
x_ft = paddle.fft.rfftn(x, axes=[2, 3])
# Multiply relevant Fourier modes
- out_ft = paddle.zeros([batchsize, self.out_channels, x.shape[-2], x.shape[-1] // 2 + 1],
- dtype=paddle.float32)
- out_ft[:, :, :self.modes1, :self.modes2] = \
- compl_mul2d(x_ft[:, :, :self.modes1, :self.modes2], self.weights1)
- out_ft[:, :, -self.modes1:, :self.modes2] = \
- compl_mul2d(x_ft[:, :, -self.modes1:, :self.modes2], self.weights2)
+ out_ft = paddle.zeros(
+ [batchsize, self.out_channels, x.shape[-2], x.shape[-1] // 2 + 1],
+ dtype=paddle.float32,
+ )
+ out_ft[:, :, : self.modes1, : self.modes2] = compl_mul2d(
+ x_ft[:, :, : self.modes1, : self.modes2], self.weights1
+ )
+ out_ft[:, :, -self.modes1 :, : self.modes2] = compl_mul2d(
+ x_ft[:, :, -self.modes1 :, : self.modes2], self.weights2
+ )
# Return to physical space
x = paddle.fft.irfftn(out_ft, s=(x.shape[-2], x.shape[-1]), axes=[2, 3])
return x
+
class SpectralConv3d(nn.Layer):
def __init__(self, in_channels, out_channels, modes1, modes2, modes3):
super(SpectralConv3d, self).__init__()
self.in_channels = in_channels
self.out_channels = out_channels
- self.modes1 = modes1 #Number of Fourier modes to multiply, at most floor(N/2) + 1
+ self.modes1 = (
+ modes1 # Number of Fourier modes to multiply, at most floor(N/2) + 1
+ )
self.modes2 = modes2
self.modes3 = modes3
- self.scale = (1 / (in_channels * out_channels))
- self.weights1 = paddle.create_parameter(shape=(in_channels, out_channels), dtype=paddle.complex64, attr=Initializer.Assign(self.scale * paddle.rand(in_channels, out_channels, self.modes1, self.modes2, self.modes3)))
- self.weights2 = paddle.create_parameter(shape=(in_channels, out_channels), dtype=paddle.complex64, attr=Initializer.Assign(self.scale * paddle.rand(in_channels, out_channels, self.modes1, self.modes2, self.modes3)))
- self.weights3 = paddle.create_parameter(shape=(in_channels, out_channels), dtype=paddle.complex64, attr=Initializer.Assign(self.scale * paddle.rand(in_channels, out_channels, self.modes1, self.modes2, self.modes3)))
- self.weights4 = paddle.create_parameter(shape=(in_channels, out_channels), dtype=paddle.complex64, attr=Initializer.Assign(self.scale * paddle.rand(in_channels, out_channels, self.modes1, self.modes2, self.modes3)))
+ self.scale = 1 / (in_channels * out_channels)
+ self.weights1 = paddle.create_parameter(
+ shape=(in_channels, out_channels),
+ dtype=paddle.complex64,
+ attr=Initializer.Assign(
+ self.scale
+ * paddle.rand(
+ in_channels, out_channels, self.modes1, self.modes2, self.modes3
+ )
+ ),
+ )
+ self.weights2 = paddle.create_parameter(
+ shape=(in_channels, out_channels),
+ dtype=paddle.complex64,
+ attr=Initializer.Assign(
+ self.scale
+ * paddle.rand(
+ in_channels, out_channels, self.modes1, self.modes2, self.modes3
+ )
+ ),
+ )
+ self.weights3 = paddle.create_parameter(
+ shape=(in_channels, out_channels),
+ dtype=paddle.complex64,
+ attr=Initializer.Assign(
+ self.scale
+ * paddle.rand(
+ in_channels, out_channels, self.modes1, self.modes2, self.modes3
+ )
+ ),
+ )
+ self.weights4 = paddle.create_parameter(
+ shape=(in_channels, out_channels),
+ dtype=paddle.complex64,
+ attr=Initializer.Assign(
+ self.scale
+ * paddle.rand(
+ in_channels, out_channels, self.modes1, self.modes2, self.modes3
+ )
+ ),
+ )
def forward(self, x):
batchsize = x.shape[0]
# Compute Fourier coeffcients up to factor of e^(- something constant)
- x_ft = paddle.fft.rfftn(x, dim=[2,3,4])
-
+ x_ft = paddle.fft.rfftn(x, dim=[2, 3, 4])
+
z_dim = min(x_ft.shape[4], self.modes3)
-
+
# Multiply relevant Fourier modes
- out_ft = paddle.zeros(batchsize, self.out_channels, x_ft.shape[2], x_ft.shape[3], self.modes3, dtype=paddle.complex64)
-
- # if x_ft.shape[4] > self.modes3, truncate; if x_ft.shape[4] < self.modes3, add zero padding
- coeff = paddle.zeros(batchsize, self.in_channels, self.modes1, self.modes2, self.modes3, dtype=paddle.complex64)
- coeff[..., :z_dim] = x_ft[:, :, :self.modes1, :self.modes2, :z_dim]
- out_ft[:, :, :self.modes1, :self.modes2, :] = compl_mul3d(coeff, self.weights1)
-
- coeff = paddle.zeros(batchsize, self.in_channels, self.modes1, self.modes2, self.modes3, dtype=paddle.complex64)
- coeff[..., :z_dim] = x_ft[:, :, -self.modes1:, :self.modes2, :z_dim]
- out_ft[:, :, -self.modes1:, :self.modes2, :] = compl_mul3d(coeff, self.weights2)
-
- coeff = paddle.zeros(batchsize, self.in_channels, self.modes1, self.modes2, self.modes3, dtype=paddle.complex64)
- coeff[..., :z_dim] = x_ft[:, :, :self.modes1, -self.modes2:, :z_dim]
- out_ft[:, :, :self.modes1, -self.modes2:, :] = compl_mul3d(coeff, self.weights3)
-
- coeff = paddle.zeros(batchsize, self.in_channels, self.modes1, self.modes2, self.modes3, dtype=paddle.complex64)
- coeff[..., :z_dim] = x_ft[:, :, -self.modes1:, -self.modes2:, :z_dim]
- out_ft[:, :, -self.modes1:, -self.modes2:, :] = compl_mul3d(coeff, self.weights4)
-
- #Return to physical space
- x = paddle.fft.irfftn(out_ft, s=(x.shape[2], x.shape[3], x.shape[4]), axes=[2,3,4])
+ out_ft = paddle.zeros(
+ batchsize,
+ self.out_channels,
+ x_ft.shape[2],
+ x_ft.shape[3],
+ self.modes3,
+ dtype=paddle.complex64,
+ )
+
+ # if x_ft.shape[4] > self.modes3, truncate; if x_ft.shape[4] < self.modes3, add zero padding
+ coeff = paddle.zeros(
+ batchsize,
+ self.in_channels,
+ self.modes1,
+ self.modes2,
+ self.modes3,
+ dtype=paddle.complex64,
+ )
+ coeff[..., :z_dim] = x_ft[:, :, : self.modes1, : self.modes2, :z_dim]
+ out_ft[:, :, : self.modes1, : self.modes2, :] = compl_mul3d(
+ coeff, self.weights1
+ )
+
+ coeff = paddle.zeros(
+ batchsize,
+ self.in_channels,
+ self.modes1,
+ self.modes2,
+ self.modes3,
+ dtype=paddle.complex64,
+ )
+ coeff[..., :z_dim] = x_ft[:, :, -self.modes1 :, : self.modes2, :z_dim]
+ out_ft[:, :, -self.modes1 :, : self.modes2, :] = compl_mul3d(
+ coeff, self.weights2
+ )
+
+ coeff = paddle.zeros(
+ batchsize,
+ self.in_channels,
+ self.modes1,
+ self.modes2,
+ self.modes3,
+ dtype=paddle.complex64,
+ )
+ coeff[..., :z_dim] = x_ft[:, :, : self.modes1, -self.modes2 :, :z_dim]
+ out_ft[:, :, : self.modes1, -self.modes2 :, :] = compl_mul3d(
+ coeff, self.weights3
+ )
+
+ coeff = paddle.zeros(
+ batchsize,
+ self.in_channels,
+ self.modes1,
+ self.modes2,
+ self.modes3,
+ dtype=paddle.complex64,
+ )
+ coeff[..., :z_dim] = x_ft[:, :, -self.modes1 :, -self.modes2 :, :z_dim]
+ out_ft[:, :, -self.modes1 :, -self.modes2 :, :] = compl_mul3d(
+ coeff, self.weights4
+ )
+
+ # Return to physical space
+ x = paddle.fft.irfftn(
+ out_ft, s=(x.shape[2], x.shape[3], x.shape[4]), axes=[2, 3, 4]
+ )
return x
+
class FourierBlock(nn.Layer):
- def __init__(self, in_channels, out_channels, modes1, modes2, modes3, act='tanh'):
+ def __init__(self, in_channels, out_channels, modes1, modes2, modes3, act="tanh"):
super(FourierBlock, self).__init__()
self.in_channel = in_channels
self.out_channel = out_channels
self.speconv = SpectralConv3d(in_channels, out_channels, modes1, modes2, modes3)
self.linear = nn.Conv1D(in_channels, out_channels, 1)
- if act == 'tanh':
+ if act == "tanh":
self.act = paddle.tanh_
- elif act == 'gelu':
+ elif act == "gelu":
self.act = nn.GELU
- elif act == 'none':
+ elif act == "none":
self.act = None
else:
- raise ValueError(f'{act} is not supported')
+ raise ValueError(f"{act} is not supported")
def forward(self, x):
- '''
+ """
input x: (batchsize, channel width, x_grid, y_grid, t_grid)
- '''
+ """
x1 = self.speconv(x)
x2 = self.linear(x.reshape(x.shape[0], self.in_channel, -1))
- out = x1 + x2.reshape(x.shape[0], self.out_channel, x.shape[2], x.shape[3], x.shape[4])
+ out = x1 + x2.reshape(
+ x.shape[0], self.out_channel, x.shape[2], x.shape[3], x.shape[4]
+ )
if self.act is not None:
out = self.act(out)
return out
-
diff --git a/jointContribution/PINO/PINO_paddle/models/fourier2d.py b/jointContribution/PINO/PINO_paddle/models/fourier2d.py
index 327369aed1..33126970a9 100644
--- a/jointContribution/PINO/PINO_paddle/models/fourier2d.py
+++ b/jointContribution/PINO/PINO_paddle/models/fourier2d.py
@@ -1,34 +1,43 @@
import paddle.nn as nn
-from .basics import SpectralConv2d
+
from .FNO_blocks import FactorizedSpectralConv2d
-from .utils import _get_act, add_padding2, remove_padding2
+from .utils import _get_act
+from .utils import add_padding2
+from .utils import remove_padding2
+
class FNO2d(nn.Layer):
- def __init__(self, modes1, modes2,
- width=64, fc_dim=128,
- layers=None,
- in_dim=3, out_dim=1,
- act='gelu',
- pad_ratio=[0., 0.]):
+ def __init__(
+ self,
+ modes1,
+ modes2,
+ width=64,
+ fc_dim=128,
+ layers=None,
+ in_dim=3,
+ out_dim=1,
+ act="gelu",
+ pad_ratio=[0.0, 0.0],
+ ):
super(FNO2d, self).__init__()
"""
Args:
- modes1: list of int, number of modes in first dimension in each layer
- modes2: list of int, number of modes in second dimension in each layer
- - width: int, optional, if layers is None, it will be initialized as [width] * [len(modes1) + 1]
+ - width: int, optional, if layers is None, it will be initialized as [width] * [len(modes1) + 1]
- in_dim: number of input channels
- out_dim: number of output channels
- act: activation function, {tanh, gelu, relu, leaky_relu}, default: gelu
- - pad_ratio: list of float, or float; portion of domain to be extended. If float, paddings are added to the right.
- If list, paddings are added to both sides. pad_ratio[0] pads left, pad_ratio[1] pads right.
+ - pad_ratio: list of float, or float; portion of domain to be extended. If float, paddings are added to the right.
+ If list, paddings are added to both sides. pad_ratio[0] pads left, pad_ratio[1] pads right.
"""
if isinstance(pad_ratio, float):
pad_ratio = [pad_ratio, pad_ratio]
else:
- assert len(pad_ratio) == 2, 'Cannot add padding in more than 2 directions'
+ assert len(pad_ratio) == 2, "Cannot add padding in more than 2 directions"
self.modes1 = modes1
self.modes2 = modes2
-
+
self.pad_ratio = pad_ratio
# input channel is 3: (a(x, y), x, y)
if layers is None:
@@ -37,13 +46,21 @@ def __init__(self, modes1, modes2,
self.layers = layers
self.fc0 = nn.Linear(in_dim, layers[0])
- self.sp_convs = nn.LayerList([FactorizedSpectralConv2d(
- in_size, out_size, mode1_num, mode2_num)
- for in_size, out_size, mode1_num, mode2_num
- in zip(self.layers, self.layers[1:], self.modes1, self.modes2)])
+ self.sp_convs = nn.LayerList(
+ [
+ FactorizedSpectralConv2d(in_size, out_size, mode1_num, mode2_num)
+ for in_size, out_size, mode1_num, mode2_num in zip(
+ self.layers, self.layers[1:], self.modes1, self.modes2
+ )
+ ]
+ )
- self.ws = nn.LayerList([nn.Conv1D(in_size, out_size, 1)
- for in_size, out_size in zip(self.layers, self.layers[1:])])
+ self.ws = nn.LayerList(
+ [
+ nn.Conv1D(in_size, out_size, 1)
+ for in_size, out_size in zip(self.layers, self.layers[1:])
+ ]
+ )
self.fc1 = nn.Linear(layers[-1], fc_dim)
self.fc2 = nn.Linear(fc_dim, layers[-1])
@@ -51,28 +68,30 @@ def __init__(self, modes1, modes2,
self.act = _get_act(act)
def forward(self, x):
- '''
+ """
Args:
- x : (batch size, x_grid, y_grid, 2)
Returns:
- x: (batch size, x_grid, y_grid, 1)
- '''
+ """
size_1, size_2 = x.shape[1], x.shape[2]
if max(self.pad_ratio) > 0:
num_pad1 = [round(i * size_1) for i in self.pad_ratio]
num_pad2 = [round(i * size_2) for i in self.pad_ratio]
else:
- num_pad1 = num_pad2 = [0.]
+ num_pad1 = num_pad2 = [0.0]
length = len(self.ws)
batchsize = x.shape[0]
x = self.fc0(x)
- x = x.transpose([0, 3, 1, 2]) # B, C, X, Y
+ x = x.transpose([0, 3, 1, 2]) # B, C, X, Y
x = add_padding2(x, num_pad1, num_pad2)
size_x, size_y = x.shape[-2], x.shape[-1]
for i, (speconv, w) in enumerate(zip(self.sp_convs, self.ws)):
x1 = speconv(x)
- x2 = w(x.reshape([batchsize, self.layers[i], -1])).reshape([batchsize, self.layers[i+1], size_x, size_y])
+ x2 = w(x.reshape([batchsize, self.layers[i], -1])).reshape(
+ [batchsize, self.layers[i + 1], size_x, size_y]
+ )
x = x1 + x2
if i != length - 1:
x = self.act(x)
diff --git a/jointContribution/PINO/PINO_paddle/models/fourier3d.py b/jointContribution/PINO/PINO_paddle/models/fourier3d.py
index 077794f645..12fb58a13b 100644
--- a/jointContribution/PINO/PINO_paddle/models/fourier3d.py
+++ b/jointContribution/PINO/PINO_paddle/models/fourier3d.py
@@ -1,18 +1,26 @@
import paddle.nn as nn
-from .basics import SpectralConv3d
+
from .FNO_blocks import FactorizedSpectralConv3d
-from .utils import add_padding, remove_padding, _get_act
+from .utils import _get_act
+from .utils import add_padding
+from .utils import remove_padding
+
class FNO3d(nn.Layer):
- def __init__(self,
- modes1, modes2, modes3,
- width=16,
- fc_dim=128,
- layers=None,
- in_dim=4, out_dim=1,
- act='gelu',
- pad_ratio=[0., 0.]):
- '''
+ def __init__(
+ self,
+ modes1,
+ modes2,
+ modes3,
+ width=16,
+ fc_dim=128,
+ layers=None,
+ in_dim=4,
+ out_dim=1,
+ act="gelu",
+ pad_ratio=[0.0, 0.0],
+ ):
+ """
Args:
modes1: list of int, first dimension maximal modes for each layer
modes2: list of int, second dimension maximal modes for each layer
@@ -23,13 +31,13 @@ def __init__(self,
out_dim: int, output dimension
act: {tanh, gelu, relu, leaky_relu}, activation function
pad_ratio: the ratio of the extended domain
- '''
+ """
super(FNO3d, self).__init__()
if isinstance(pad_ratio, float):
pad_ratio = [pad_ratio, pad_ratio]
else:
- assert len(pad_ratio) == 2, 'Cannot add padding in more than 2 directions.'
+ assert len(pad_ratio) == 2, "Cannot add padding in more than 2 directions."
self.pad_ratio = pad_ratio
self.modes1 = modes1
@@ -43,35 +51,45 @@ def __init__(self,
self.layers = layers
self.fc0 = nn.Linear(in_dim, layers[0])
- self.sp_convs = nn.LayerList([FactorizedSpectralConv3d(
- in_size, out_size, mode1_num, mode2_num, mode3_num)
- for in_size, out_size, mode1_num, mode2_num, mode3_num
- in zip(self.layers, self.layers[1:], self.modes1, self.modes2, self.modes3)])
+ self.sp_convs = nn.LayerList(
+ [
+ FactorizedSpectralConv3d(
+ in_size, out_size, mode1_num, mode2_num, mode3_num
+ )
+ for in_size, out_size, mode1_num, mode2_num, mode3_num in zip(
+ self.layers, self.layers[1:], self.modes1, self.modes2, self.modes3
+ )
+ ]
+ )
- self.ws = nn.LayerList([nn.Conv1D(in_size, out_size, 1)
- for in_size, out_size in zip(self.layers, self.layers[1:])])
+ self.ws = nn.LayerList(
+ [
+ nn.Conv1D(in_size, out_size, 1)
+ for in_size, out_size in zip(self.layers, self.layers[1:])
+ ]
+ )
self.fc1 = nn.Linear(layers[-1], fc_dim)
self.fc2 = nn.Linear(fc_dim, out_dim)
self.act = _get_act(act)
def forward(self, x):
- '''
+ """
Args:
x: (batchsize, x_grid, y_grid, t_grid, 3)
Returns:
u: (batchsize, x_grid, y_grid, t_grid, 1)
- '''
+ """
size_z = x.shape[-2]
if max(self.pad_ratio) > 0:
num_pad = [round(size_z * i) for i in self.pad_ratio]
else:
- num_pad = [0., 0.]
+ num_pad = [0.0, 0.0]
length = len(self.ws)
batchsize = x.shape[0]
-
+
x = self.fc0(x)
x = x.transpose([0, 4, 1, 2, 3])
x = add_padding(x, num_pad=num_pad)
@@ -79,7 +97,9 @@ def forward(self, x):
for i, (speconv, w) in enumerate(zip(self.sp_convs, self.ws)):
x1 = speconv(x)
- x2 = w(x.reshape([batchsize, self.layers[i], -1])).reshape([batchsize, self.layers[i+1], size_x, size_y, size_z])
+ x2 = w(x.reshape([batchsize, self.layers[i], -1])).reshape(
+ [batchsize, self.layers[i + 1], size_x, size_y, size_z]
+ )
x = x1 + x2
if i != length - 1:
x = self.act(x)
@@ -88,4 +108,4 @@ def forward(self, x):
x = self.fc1(x)
x = self.act(x)
x = self.fc2(x)
- return x
\ No newline at end of file
+ return x
diff --git a/jointContribution/PINO/PINO_paddle/models/utils.py b/jointContribution/PINO/PINO_paddle/models/utils.py
index 0302c5ab3c..2ae65ef56c 100644
--- a/jointContribution/PINO/PINO_paddle/models/utils.py
+++ b/jointContribution/PINO/PINO_paddle/models/utils.py
@@ -1,45 +1,51 @@
import paddle.nn.functional as F
+
def add_padding(x, num_pad):
if max(num_pad) > 0:
- res = F.pad(x, (num_pad[0], num_pad[1]), 'constant', 0)
+ res = F.pad(x, (num_pad[0], num_pad[1]), "constant", 0)
else:
res = x
return res
+
def add_padding2(x, num_pad1, num_pad2):
if max(num_pad1) > 0 or max(num_pad2) > 0:
- res = F.pad(x, (num_pad2[0], num_pad2[1], num_pad1[0], num_pad1[1]), 'constant', 0.)
+ res = F.pad(
+ x, (num_pad2[0], num_pad2[1], num_pad1[0], num_pad1[1]), "constant", 0.0
+ )
else:
res = x
return res
+
def remove_padding(x, num_pad):
if max(num_pad) > 0:
- res = x[..., num_pad[0]:-num_pad[1]]
+ res = x[..., num_pad[0] : -num_pad[1]]
else:
res = x
return res
+
def remove_padding2(x, num_pad1, num_pad2):
if max(num_pad1) > 0 or max(num_pad2) > 0:
- res = x[..., num_pad1[0]:-num_pad1[1], num_pad2[0]:-num_pad2[1]]
+ res = x[..., num_pad1[0] : -num_pad1[1], num_pad2[0] : -num_pad2[1]]
else:
res = x
return res
+
def _get_act(act):
- if act == 'tanh':
+ if act == "tanh":
func = F.tanh
- elif act == 'gelu':
+ elif act == "gelu":
func = F.gelu
- elif act == 'relu':
+ elif act == "relu":
func = F.relu_
- elif act == 'elu':
+ elif act == "elu":
func = F.elu_
- elif act == 'leaky_relu':
+ elif act == "leaky_relu":
func = F.leaky_relu
else:
- raise ValueError(f'{act} is not supported')
+ raise ValueError(f"{act} is not supported")
return func
-
diff --git a/jointContribution/PINO/PINO_paddle/prepare_data.py b/jointContribution/PINO/PINO_paddle/prepare_data.py
index 74a14694e1..2dc4349a81 100644
--- a/jointContribution/PINO/PINO_paddle/prepare_data.py
+++ b/jointContribution/PINO/PINO_paddle/prepare_data.py
@@ -1,33 +1,35 @@
import numpy as np
-import matplotlib.pyplot as plt
+
def shuffle_data(datapath):
data = np.load(datapath)
rng = np.random.default_rng(123)
rng.shuffle(data, axis=0)
- savepath = datapath.replace('.npy', '-shuffle.npy')
+ savepath = datapath.replace(".npy", "-shuffle.npy")
np.save(savepath, data)
+
def test_data(datapath):
- raw = np.load(datapath, mmap_mode='r')
+ raw = np.load(datapath, mmap_mode="r")
print(raw[0, 0, 0, 0:10])
- newpath = datapath.replace('.npy', '-shuffle.npy')
- new = np.load(newpath, mmap_mode='r')
+ newpath = datapath.replace(".npy", "-shuffle.npy")
+ new = np.load(newpath, mmap_mode="r")
print(new[0, 0, 0, 0:10])
+
def get_slice(datapath):
- raw = np.load(datapath, mmap_mode='r')
+ raw = np.load(datapath, mmap_mode="r")
data = raw[-10:]
print(data.shape)
- savepath = 'data/Re500-5x513x256x256.npy'
+ savepath = "data/Re500-5x513x256x256.npy"
np.save(savepath, data)
+
def plot_test(datapath):
- duration = 0.125
- raw = np.load(datapath, mmap_mode='r')
-
+ np.load(datapath, mmap_mode="r")
+
-if __name__ == '__main__':
- datapath = '/raid/hongkai/NS-Re500_T300_id0-shuffle.npy'
- get_slice(datapath)
\ No newline at end of file
+if __name__ == "__main__":
+ datapath = "/raid/hongkai/NS-Re500_T300_id0-shuffle.npy"
+ get_slice(datapath)
diff --git a/jointContribution/PINO/PINO_paddle/solver/random_fields.py b/jointContribution/PINO/PINO_paddle/solver/random_fields.py
index 448567db3e..458c1a0e72 100644
--- a/jointContribution/PINO/PINO_paddle/solver/random_fields.py
+++ b/jointContribution/PINO/PINO_paddle/solver/random_fields.py
@@ -1,58 +1,102 @@
+import math
+
import paddle
-import math
class GaussianRF(object):
- def __init__(self, dim, size, length=1.0, alpha=2.0, tau=3.0, sigma=None, boundary="periodic", constant_eig=False):
+ def __init__(
+ self,
+ dim,
+ size,
+ length=1.0,
+ alpha=2.0,
+ tau=3.0,
+ sigma=None,
+ boundary="periodic",
+ constant_eig=False,
+ ):
self.dim = dim
if sigma is None:
- sigma = tau**(0.5*(2*alpha - self.dim))
+ sigma = tau ** (0.5 * (2 * alpha - self.dim))
- k_max = size//2
+ k_max = size // 2
- const = (4*(math.pi**2))/(length**2)
+ const = (4 * (math.pi**2)) / (length**2)
if dim == 1:
- k = paddle.concat((paddle.arange(start=0, end=k_max, step=1), \
- paddle.arange(start=-k_max, end=0, step=1)), 0)
-
- self.sqrt_eig = size*math.sqrt(2.0)*sigma*((const*(k**2) + tau**2)**(-alpha/2.0))
+ k = paddle.concat(
+ (
+ paddle.arange(start=0, end=k_max, step=1),
+ paddle.arange(start=-k_max, end=0, step=1),
+ ),
+ 0,
+ )
+
+ self.sqrt_eig = (
+ size
+ * math.sqrt(2.0)
+ * sigma
+ * ((const * (k**2) + tau**2) ** (-alpha / 2.0))
+ )
if constant_eig:
- self.sqrt_eig[0] = size*sigma*(tau**(-alpha))
+ self.sqrt_eig[0] = size * sigma * (tau ** (-alpha))
else:
self.sqrt_eig[0] = 0.0
elif dim == 2:
- wavenumers = paddle.concat((paddle.arange(start=0, end=k_max, step=1), \
- paddle.arange(start=-k_max, end=0, step=1)), 0).repeat(size,1)
-
- k_x = wavenumers.transpose(0,1)
+ wavenumers = paddle.concat(
+ (
+ paddle.arange(start=0, end=k_max, step=1),
+ paddle.arange(start=-k_max, end=0, step=1),
+ ),
+ 0,
+ ).repeat(size, 1)
+
+ k_x = wavenumers.transpose(0, 1)
k_y = wavenumers
- self.sqrt_eig = (size**2)*math.sqrt(2.0)*sigma*((const*(k_x**2 + k_y**2) + tau**2)**(-alpha/2.0))
+ self.sqrt_eig = (
+ (size**2)
+ * math.sqrt(2.0)
+ * sigma
+ * ((const * (k_x**2 + k_y**2) + tau**2) ** (-alpha / 2.0))
+ )
if constant_eig:
- self.sqrt_eig[0,0] = (size**2)*sigma*(tau**(-alpha))
+ self.sqrt_eig[0, 0] = (size**2) * sigma * (tau ** (-alpha))
else:
- self.sqrt_eig[0,0] = 0.0
+ self.sqrt_eig[0, 0] = 0.0
elif dim == 3:
- wavenumers = paddle.concat((paddle.arange(start=0, end=k_max, step=1), \
- paddle.arange(start=-k_max, end=0, step=1)), 0).repeat(size,size,1)
-
- k_x = wavenumers.transpose(1,2)
+ wavenumers = paddle.concat(
+ (
+ paddle.arange(start=0, end=k_max, step=1),
+ paddle.arange(start=-k_max, end=0, step=1),
+ ),
+ 0,
+ ).repeat(size, size, 1)
+
+ k_x = wavenumers.transpose(1, 2)
k_y = wavenumers
- k_z = wavenumers.transpose(0,2)
-
- self.sqrt_eig = (size**3)*math.sqrt(2.0)*sigma*((const*(k_x**2 + k_y**2 + k_z**2) + tau**2)**(-alpha/2.0))
+ k_z = wavenumers.transpose(0, 2)
+
+ self.sqrt_eig = (
+ (size**3)
+ * math.sqrt(2.0)
+ * sigma
+ * (
+ (const * (k_x**2 + k_y**2 + k_z**2) + tau**2)
+ ** (-alpha / 2.0)
+ )
+ )
if constant_eig:
- self.sqrt_eig[0,0,0] = (size**3)*sigma*(tau**(-alpha))
+ self.sqrt_eig[0, 0, 0] = (size**3) * sigma * (tau ** (-alpha))
else:
- self.sqrt_eig[0,0,0] = 0.0
+ self.sqrt_eig[0, 0, 0] = 0.0
self.size = []
for j in range(self.dim):
@@ -63,14 +107,26 @@ def __init__(self, dim, size, length=1.0, alpha=2.0, tau=3.0, sigma=None, bounda
def sample(self, N):
coeff = paddle.randn(N, *self.size, dtype=paddle.float32)
- coeff = self.sqrt_eig*coeff
+ coeff = self.sqrt_eig * coeff
u = paddle.fft.irfftn(coeff, self.size, norm="backward")
return u
-class GaussianRF2d(object):
- def __init__(self, s1, s2, L1=2*math.pi, L2=2*math.pi, alpha=2.0, tau=3.0, sigma=None, mean=None, boundary="periodic", dtype=paddle.float64):
+class GaussianRF2d(object):
+ def __init__(
+ self,
+ s1,
+ s2,
+ L1=2 * math.pi,
+ L2=2 * math.pi,
+ alpha=2.0,
+ tau=3.0,
+ sigma=None,
+ mean=None,
+ boundary="periodic",
+ dtype=paddle.float64,
+ ):
self.s1 = s1
self.s2 = s2
@@ -80,34 +136,44 @@ def __init__(self, s1, s2, L1=2*math.pi, L2=2*math.pi, alpha=2.0, tau=3.0, sigma
self.dtype = dtype
if sigma is None:
- self.sigma = tau**(0.5*(2*alpha - 2.0))
+ self.sigma = tau ** (0.5 * (2 * alpha - 2.0))
else:
self.sigma = sigma
- const1 = (4*(math.pi**2))/(L1**2)
- const2 = (4*(math.pi**2))/(L2**2)
+ const1 = (4 * (math.pi**2)) / (L1**2)
+ const2 = (4 * (math.pi**2)) / (L2**2)
- freq_list1 = paddle.concat((paddle.arange(start=0, end=s1//2, step=1),\
- paddle.arange(start=-s1//2, end=0, step=1)), 0)
- k1 = freq_list1.reshape([-1,1]).repeat([1, s2//2 + 1]).type(dtype)
+ freq_list1 = paddle.concat(
+ (
+ paddle.arange(start=0, end=s1 // 2, step=1),
+ paddle.arange(start=-s1 // 2, end=0, step=1),
+ ),
+ 0,
+ )
+ k1 = freq_list1.reshape([-1, 1]).repeat([1, s2 // 2 + 1]).type(dtype)
- freq_list2 = paddle.arange(start=0, end=s2//2 + 1, step=1)
+ freq_list2 = paddle.arange(start=0, end=s2 // 2 + 1, step=1)
- k2 = freq_list2.view(1,-1).repeat(s1, 1).type(dtype)
+ k2 = freq_list2.view(1, -1).repeat(s1, 1).type(dtype)
- self.sqrt_eig = s1*s2*self.sigma*((const1*k1**2 + const2*k2**2 + tau**2)**(-alpha/2.0))
- self.sqrt_eig[0,0] = 0.0
+ self.sqrt_eig = (
+ s1
+ * s2
+ * self.sigma
+ * ((const1 * k1**2 + const2 * k2**2 + tau**2) ** (-alpha / 2.0))
+ )
+ self.sqrt_eig[0, 0] = 0.0
def sample(self, N, xi=None):
if xi is None:
- xi = paddle.randn(N, self.s1, self.s2//2 + 1, 2, dtype=self.dtype)
-
- xi[...,0] = self.sqrt_eig*xi [...,0]
- xi[...,1] = self.sqrt_eig*xi [...,1]
-
+ xi = paddle.randn(N, self.s1, self.s2 // 2 + 1, 2, dtype=self.dtype)
+
+ xi[..., 0] = self.sqrt_eig * xi[..., 0]
+ xi[..., 1] = self.sqrt_eig * xi[..., 1]
+
u = paddle.fft.irfft2(paddle.reshape(xi), s=(self.s1, self.s2))
if self.mean is not None:
u += self.mean
-
- return u
\ No newline at end of file
+
+ return u
diff --git a/jointContribution/PINO/PINO_paddle/train_burgers.py b/jointContribution/PINO/PINO_paddle/train_burgers.py
index 7b5d2d614b..a309349f94 100644
--- a/jointContribution/PINO/PINO_paddle/train_burgers.py
+++ b/jointContribution/PINO/PINO_paddle/train_burgers.py
@@ -1,85 +1,113 @@
-import yaml
from argparse import ArgumentParser
+
import paddle
-from paddle.optimizer.lr import MultiStepDecay
-from paddle.optimizer import Adam
+import yaml
from models import FNO2d
+from paddle.optimizer import Adam
+from paddle.optimizer.lr import MultiStepDecay
from train_utils.datasets import BurgersLoader
-from train_utils.train_2d import train_2d_burger
from train_utils.eval_2d import eval_burgers
+from train_utils.train_2d import train_2d_burger
+
def run(args, config):
- data_config = config['data']
- dataset = BurgersLoader(data_config['datapath'],
- nx=data_config['nx'], nt=data_config['nt'],
- sub=data_config['sub'], sub_t=data_config['sub_t'], new=True)
- train_loader = dataset.make_loader(n_sample=data_config['n_sample'],
- batch_size=config['train']['batchsize'],
- start=data_config['offset'])
+ data_config = config["data"]
+ dataset = BurgersLoader(
+ data_config["datapath"],
+ nx=data_config["nx"],
+ nt=data_config["nt"],
+ sub=data_config["sub"],
+ sub_t=data_config["sub_t"],
+ new=True,
+ )
+ train_loader = dataset.make_loader(
+ n_sample=data_config["n_sample"],
+ batch_size=config["train"]["batchsize"],
+ start=data_config["offset"],
+ )
- model = FNO2d(modes1=config['model']['modes1'],
- modes2=config['model']['modes2'],
- fc_dim=config['model']['fc_dim'],
- layers=config['model']['layers'],
- act=config['model']['act'])
- param_state_dict = paddle.load('init_param/init_burgers.pdparams')
+ model = FNO2d(
+ modes1=config["model"]["modes1"],
+ modes2=config["model"]["modes2"],
+ fc_dim=config["model"]["fc_dim"],
+ layers=config["model"]["layers"],
+ act=config["model"]["act"],
+ )
+ param_state_dict = paddle.load("init_param/init_burgers.pdparams")
model.set_dict(param_state_dict)
# Load from checkpoint
- if 'ckpt' in config['train']:
- ckpt_path = config['train']['ckpt']
+ if "ckpt" in config["train"]:
+ ckpt_path = config["train"]["ckpt"]
ckpt = paddle.load(ckpt_path)
- model.load_state_dict(ckpt['model'])
- print('Weights loaded from %s' % ckpt_path)
+ model.load_state_dict(ckpt["model"])
+ print("Weights loaded from %s" % ckpt_path)
- scheduler = MultiStepDecay(learning_rate=config['train']['base_lr'],
- milestones=config['train']['milestones'],
- gamma=config['train']['scheduler_gamma'])
+ scheduler = MultiStepDecay(
+ learning_rate=config["train"]["base_lr"],
+ milestones=config["train"]["milestones"],
+ gamma=config["train"]["scheduler_gamma"],
+ )
optimizer = Adam(learning_rate=scheduler, parameters=model.parameters())
-
- train_2d_burger(model,
- train_loader,
- dataset.v,
- optimizer,
- scheduler,
- config,
- rank=0,
- log=args.log,
- project=config['log']['project'],
- group=config['log']['group'])
+
+ train_2d_burger(
+ model,
+ train_loader,
+ dataset.v,
+ optimizer,
+ scheduler,
+ config,
+ rank=0,
+ log=args.log,
+ project=config["log"]["project"],
+ group=config["log"]["group"],
+ )
+
def test(config):
- data_config = config['data']
- dataset = BurgersLoader(data_config['datapath'],
- nx=data_config['nx'], nt=data_config['nt'],
- sub=data_config['sub'], sub_t=data_config['sub_t'], new=True)
- dataloader = dataset.make_loader(n_sample=data_config['n_sample'],
- batch_size=config['test']['batchsize'],
- start=data_config['offset'])
+ data_config = config["data"]
+ dataset = BurgersLoader(
+ data_config["datapath"],
+ nx=data_config["nx"],
+ nt=data_config["nt"],
+ sub=data_config["sub"],
+ sub_t=data_config["sub_t"],
+ new=True,
+ )
+ dataloader = dataset.make_loader(
+ n_sample=data_config["n_sample"],
+ batch_size=config["test"]["batchsize"],
+ start=data_config["offset"],
+ )
- model = FNO2d(modes1=config['model']['modes1'],
- modes2=config['model']['modes2'],
- fc_dim=config['model']['fc_dim'],
- layers=config['model']['layers'],
- act=config['model']['act'])
+ model = FNO2d(
+ modes1=config["model"]["modes1"],
+ modes2=config["model"]["modes2"],
+ fc_dim=config["model"]["fc_dim"],
+ layers=config["model"]["layers"],
+ act=config["model"]["act"],
+ )
# Load from checkpoint
- if 'ckpt' in config['test']:
- ckpt_path = config['test']['ckpt']
+ if "ckpt" in config["test"]:
+ ckpt_path = config["test"]["ckpt"]
ckpt = paddle.load(ckpt_path)
- model.load_state_dict(ckpt['model'])
- print('Weights loaded from %s' % ckpt_path)
+ model.load_state_dict(ckpt["model"])
+ print("Weights loaded from %s" % ckpt_path)
eval_burgers(model, dataloader, dataset.v, config)
-if __name__ == '__main__':
- parser = ArgumentParser(description='Basic paser')
- parser.add_argument('--config_path', type=str, help='Path to the configuration file')
- parser.add_argument('--log', action='store_true', help='Turn on the wandb')
- parser.add_argument('--mode', type=str, help='train or test')
+
+if __name__ == "__main__":
+ parser = ArgumentParser(description="Basic paser")
+ parser.add_argument(
+ "--config_path", type=str, help="Path to the configuration file"
+ )
+ parser.add_argument("--log", action="store_true", help="Turn on the wandb")
+ parser.add_argument("--mode", type=str, help="train or test")
args = parser.parse_args()
config_file = args.config_path
- with open(config_file, 'r') as stream:
+ with open(config_file, "r") as stream:
config = yaml.load(stream, yaml.FullLoader)
- if args.mode == 'train':
+ if args.mode == "train":
run(args, config)
else:
test(config)
diff --git a/jointContribution/PINO/PINO_paddle/train_operator.py b/jointContribution/PINO/PINO_paddle/train_operator.py
index 5f9fd6ae1c..0a95277336 100644
--- a/jointContribution/PINO/PINO_paddle/train_operator.py
+++ b/jointContribution/PINO/PINO_paddle/train_operator.py
@@ -1,123 +1,164 @@
-import yaml
-from argparse import ArgumentParser
import math
+from argparse import ArgumentParser
import paddle
+import yaml
+from models import FNO2d
+from models import FNO3d
from paddle.io import DataLoader
from paddle.optimizer.lr import MultiStepDecay
-
from solver.random_fields import GaussianRF
from train_utils import Adam
-from train_utils.datasets import NSLoader, online_loader, DarcyFlow, DarcyCombo
-from train_utils.train_3d import mixed_train
+from train_utils.datasets import DarcyCombo
+from train_utils.datasets import NSLoader
+from train_utils.datasets import online_loader
from train_utils.train_2d import train_2d_operator
-from models import FNO3d, FNO2d
+from train_utils.train_3d import mixed_train
+
def train_3d(args, config):
- data_config = config['data']
+ data_config = config["data"]
# prepare dataloader for training with data
- if 'datapath2' in data_config:
- loader = NSLoader(datapath1=data_config['datapath'], datapath2=data_config['datapath2'],
- nx=data_config['nx'], nt=data_config['nt'],
- sub=data_config['sub'], sub_t=data_config['sub_t'],
- N=data_config['total_num'],
- t_interval=data_config['time_interval'])
+ if "datapath2" in data_config:
+ loader = NSLoader(
+ datapath1=data_config["datapath"],
+ datapath2=data_config["datapath2"],
+ nx=data_config["nx"],
+ nt=data_config["nt"],
+ sub=data_config["sub"],
+ sub_t=data_config["sub_t"],
+ N=data_config["total_num"],
+ t_interval=data_config["time_interval"],
+ )
else:
- loader = NSLoader(datapath1=data_config['datapath'],
- nx=data_config['nx'], nt=data_config['nt'],
- sub=data_config['sub'], sub_t=data_config['sub_t'],
- N=data_config['total_num'],
- t_interval=data_config['time_interval'])
+ loader = NSLoader(
+ datapath1=data_config["datapath"],
+ nx=data_config["nx"],
+ nt=data_config["nt"],
+ sub=data_config["sub"],
+ sub_t=data_config["sub_t"],
+ N=data_config["total_num"],
+ t_interval=data_config["time_interval"],
+ )
- train_loader = loader.make_loader(data_config['n_sample'],
- batch_size=config['train']['batchsize'],
- start=data_config['offset'],
- train=data_config['shuffle'])
+ train_loader = loader.make_loader(
+ data_config["n_sample"],
+ batch_size=config["train"]["batchsize"],
+ start=data_config["offset"],
+ train=data_config["shuffle"],
+ )
# prepare dataloader for training with only equations
- gr_sampler = GaussianRF(2, data_config['S2'], 2 * math.pi, alpha=2.5, tau=7)
- a_loader = online_loader(gr_sampler,
- S=data_config['S2'],
- T=data_config['T2'],
- time_scale=data_config['time_interval'],
- batchsize=config['train']['batchsize'])
+ gr_sampler = GaussianRF(2, data_config["S2"], 2 * math.pi, alpha=2.5, tau=7)
+ a_loader = online_loader(
+ gr_sampler,
+ S=data_config["S2"],
+ T=data_config["T2"],
+ time_scale=data_config["time_interval"],
+ batchsize=config["train"]["batchsize"],
+ )
# create model
- model = FNO3d(modes1=config['model']['modes1'],
- modes2=config['model']['modes2'],
- modes3=config['model']['modes3'],
- fc_dim=config['model']['fc_dim'],
- layers=config['model']['layers'],
- act=config['model']['act'])
+ model = FNO3d(
+ modes1=config["model"]["modes1"],
+ modes2=config["model"]["modes2"],
+ modes3=config["model"]["modes3"],
+ fc_dim=config["model"]["fc_dim"],
+ layers=config["model"]["layers"],
+ act=config["model"]["act"],
+ )
# Load from checkpoint
- if 'ckpt' in config['train']:
- ckpt_path = config['train']['ckpt']
+ if "ckpt" in config["train"]:
+ ckpt_path = config["train"]["ckpt"]
ckpt = paddle.load(ckpt_path)
- model.load_state_dict(ckpt['model'])
- print('Weights loaded from %s' % ckpt_path)
+ model.load_state_dict(ckpt["model"])
+ print("Weights loaded from %s" % ckpt_path)
# create optimizer and learning rate scheduler
- scheduler = MultiStepDecay(learning_rate=config['train']['base_lr'],
- milestones=config['train']['milestones'],
- gamma=config['train']['scheduler_gamma'])
+ scheduler = MultiStepDecay(
+ learning_rate=config["train"]["base_lr"],
+ milestones=config["train"]["milestones"],
+ gamma=config["train"]["scheduler_gamma"],
+ )
optimizer = Adam(learning_rate=scheduler, parameters=model.parameters())
- mixed_train(model,
- train_loader,
- loader.S, loader.T,
- a_loader,
- data_config['S2'], data_config['T2'],
- optimizer,
- scheduler,
- config,
- log=args.log,
- project=config['log']['project'],
- group=config['log']['group'])
+ mixed_train(
+ model,
+ train_loader,
+ loader.S,
+ loader.T,
+ a_loader,
+ data_config["S2"],
+ data_config["T2"],
+ optimizer,
+ scheduler,
+ config,
+ log=args.log,
+ project=config["log"]["project"],
+ group=config["log"]["group"],
+ )
+
def train_2d(args, config):
- data_config = config['data']
+ data_config = config["data"]
- dataset = DarcyCombo(datapath=data_config['datapath'],
- nx=data_config['nx'],
- sub=data_config['sub'],
- pde_sub=data_config['pde_sub'],
- num=data_config['n_sample'],
- offset=data_config['offset'])
- train_loader = DataLoader(dataset, batch_size=config['train']['batchsize'], shuffle=True)
- model = FNO2d(modes1=config['model']['modes1'],
- modes2=config['model']['modes2'],
- fc_dim=config['model']['fc_dim'],
- layers=config['model']['layers'],
- act=config['model']['act'],
- pad_ratio=config['model']['pad_ratio'])
+ dataset = DarcyCombo(
+ datapath=data_config["datapath"],
+ nx=data_config["nx"],
+ sub=data_config["sub"],
+ pde_sub=data_config["pde_sub"],
+ num=data_config["n_sample"],
+ offset=data_config["offset"],
+ )
+ train_loader = DataLoader(
+ dataset, batch_size=config["train"]["batchsize"], shuffle=True
+ )
+ model = FNO2d(
+ modes1=config["model"]["modes1"],
+ modes2=config["model"]["modes2"],
+ fc_dim=config["model"]["fc_dim"],
+ layers=config["model"]["layers"],
+ act=config["model"]["act"],
+ pad_ratio=config["model"]["pad_ratio"],
+ )
# Load from checkpoint
- if 'ckpt' in config['train']:
- ckpt_path = config['train']['ckpt']
+ if "ckpt" in config["train"]:
+ ckpt_path = config["train"]["ckpt"]
ckpt = paddle.load(ckpt_path)
- model.load_state_dict(ckpt['model'])
- print('Weights loaded from %s' % ckpt_path)
+ model.load_state_dict(ckpt["model"])
+ print("Weights loaded from %s" % ckpt_path)
- scheduler = MultiStepDecay(learning_rate=config['train']['base_lr'],
- milestones=config['train']['milestones'],
- gamma=config['train']['scheduler_gamma'])
+ scheduler = MultiStepDecay(
+ learning_rate=config["train"]["base_lr"],
+ milestones=config["train"]["milestones"],
+ gamma=config["train"]["scheduler_gamma"],
+ )
optimizer = Adam(learning_rate=scheduler, parameters=model.parameters())
- train_2d_operator(model,
- train_loader,
- optimizer, scheduler,
- config, rank=0, log=args.log,
- project=config['log']['project'],
- group=config['log']['group'])
+ train_2d_operator(
+ model,
+ train_loader,
+ optimizer,
+ scheduler,
+ config,
+ rank=0,
+ log=args.log,
+ project=config["log"]["project"],
+ group=config["log"]["group"],
+ )
+
-if __name__ == '__main__':
+if __name__ == "__main__":
# parse options
- parser = ArgumentParser(description='Basic paser')
- parser.add_argument('--config_path', type=str, help='Path to the configuration file')
- parser.add_argument('--log', action='store_true', help='Turn on the wandb')
+ parser = ArgumentParser(description="Basic paser")
+ parser.add_argument(
+ "--config_path", type=str, help="Path to the configuration file"
+ )
+ parser.add_argument("--log", action="store_true", help="Turn on the wandb")
args = parser.parse_args()
config_file = args.config_path
- with open(config_file, 'r') as stream:
+ with open(config_file, "r") as stream:
config = yaml.load(stream, yaml.FullLoader)
- if 'name' in config['data'] and config['data']['name'] == 'Darcy':
+ if "name" in config["data"] and config["data"]["name"] == "Darcy":
train_2d(args, config)
else:
train_3d(args, config)
diff --git a/jointContribution/PINO/PINO_paddle/train_pino.py b/jointContribution/PINO/PINO_paddle/train_pino.py
index 828ae24702..7fbf25af53 100644
--- a/jointContribution/PINO/PINO_paddle/train_pino.py
+++ b/jointContribution/PINO/PINO_paddle/train_pino.py
@@ -1,17 +1,25 @@
import os
-import yaml
import random
from argparse import ArgumentParser
-from tqdm import tqdm
+
import numpy as np
import paddle
-from paddle.optimizer import Adam
+import yaml
+from models import FNO3d
from paddle.io import DataLoader
+from paddle.optimizer import Adam
from paddle.optimizer.lr import MultiStepDecay
-from models import FNO3d
-from train_utils.losses import LpLoss, PINO_loss3d, get_forcing
-from train_utils.datasets import KFDataset, KFaDataset, sample_data
-from train_utils.utils import save_ckpt, count_params, dict2str
+from tqdm import tqdm
+from train_utils.datasets import KFaDataset
+from train_utils.datasets import KFDataset
+from train_utils.datasets import sample_data
+from train_utils.losses import LpLoss
+from train_utils.losses import PINO_loss3d
+from train_utils.losses import get_forcing
+from train_utils.utils import count_params
+from train_utils.utils import dict2str
+from train_utils.utils import save_ckpt
+
@paddle.no_grad()
def eval_ns(model, val_loader, criterion):
@@ -29,34 +37,38 @@ def eval_ns(model, val_loader, criterion):
std_err = np.std(val_err, ddof=1) / np.sqrt(N)
return avg_err, std_err
-def train_ns(model,
- train_u_loader, # training data
- train_a_loader, # initial conditions
- val_loader, # validation data
- optimizer,
- scheduler,
- config, args):
- start_iter = config['train']['start_iter']
- v = 1/ config['data']['Re']
- t_duration = config['data']['t_duration']
- save_step = config['train']['save_step']
- eval_step = config['train']['eval_step']
-
- ic_weight = config['train']['ic_loss']
- f_weight = config['train']['f_loss']
- xy_weight = config['train']['xy_loss']
+
+def train_ns(
+ model,
+ train_u_loader, # training data
+ train_a_loader, # initial conditions
+ val_loader, # validation data
+ optimizer,
+ scheduler,
+ config,
+ args,
+):
+ start_iter = config["train"]["start_iter"]
+ v = 1 / config["data"]["Re"]
+ t_duration = config["data"]["t_duration"]
+ save_step = config["train"]["save_step"]
+ eval_step = config["train"]["eval_step"]
+
+ ic_weight = config["train"]["ic_loss"]
+ f_weight = config["train"]["f_loss"]
+ xy_weight = config["train"]["xy_loss"]
# set up directory
- base_dir = os.path.join('exp', config['log']['logdir'])
- ckpt_dir = os.path.join(base_dir, 'ckpts')
+ base_dir = os.path.join("exp", config["log"]["logdir"])
+ ckpt_dir = os.path.join(base_dir, "ckpts")
os.makedirs(ckpt_dir, exist_ok=True)
# loss fn
lploss = LpLoss(size_average=True)
-
- S = config['data']['pde_res'][0]
+
+ S = config["data"]["pde_res"][0]
forcing = get_forcing(S)
-
- pbar = range(start_iter, config['train']['num_iter'])
+
+ pbar = range(start_iter, config["train"]["num_iter"])
if args.tqdm:
pbar = tqdm(pbar, dynamic_ncols=True, smoothing=0.2)
@@ -75,18 +87,18 @@ def train_ns(model,
out = model(a_in)
data_loss = lploss(out, u)
else:
- data_loss = paddle.zeros(1, dtype='float32')
+ data_loss = paddle.zeros(1, dtype="float32")
if f_weight != 0.0:
# pde loss
a = next(a_loader)
a = a
out = model(a)
-
- u0 = a[:, :, :, 0, -1]
+
+ u0 = a[:, :, :, 0, -1]
loss_ic, loss_f = PINO_loss3d(out, u0, forcing, v, t_duration)
- log_dict['IC'] = loss_ic.item()
- log_dict['PDE'] = loss_f.item()
+ log_dict["IC"] = loss_ic.item()
+ log_dict["PDE"] = loss_f.item()
else:
loss_ic = loss_f = 0.0
loss = data_loss * xy_weight + loss_f * f_weight + loss_ic * ic_weight
@@ -95,125 +107,131 @@ def train_ns(model,
optimizer.step()
scheduler.step()
- log_dict['train loss'] = loss.item()
- log_dict['data'] = data_loss.item()
+ log_dict["train loss"] = loss.item()
+ log_dict["data"] = data_loss.item()
if e % eval_step == 0:
eval_err, std_err = eval_ns(model, val_loader, lploss)
- log_dict['val error'] = eval_err
-
+ log_dict["val error"] = eval_err
+
if args.tqdm:
logstr = dict2str(log_dict)
- pbar.set_description(
- (
- logstr
- )
- )
+ pbar.set_description((logstr))
if e % save_step == 0 and e > 0:
- ckpt_path = os.path.join(ckpt_dir, f'model-{e}.pt')
+ ckpt_path = os.path.join(ckpt_dir, f"model-{e}.pt")
save_ckpt(ckpt_path, model, optimizer, scheduler)
+
def subprocess(args):
- with open(args.config, 'r') as f:
+ with open(args.config, "r") as f:
config = yaml.load(f, yaml.FullLoader)
# set random seed
- config['seed'] = args.seed
+ config["seed"] = args.seed
seed = args.seed
paddle.seed(seed)
random.seed(seed)
if paddle.cuda.is_available():
paddle.cuda.manual_seed_all(seed)
- # create model
- model = FNO3d(modes1=config['model']['modes1'],
- modes2=config['model']['modes2'],
- modes3=config['model']['modes3'],
- fc_dim=config['model']['fc_dim'],
- layers=config['model']['layers'],
- act=config['model']['act'],
- pad_ratio=config['model']['pad_ratio'])
+ # create model
+ model = FNO3d(
+ modes1=config["model"]["modes1"],
+ modes2=config["model"]["modes2"],
+ modes3=config["model"]["modes3"],
+ fc_dim=config["model"]["fc_dim"],
+ layers=config["model"]["layers"],
+ act=config["model"]["act"],
+ pad_ratio=config["model"]["pad_ratio"],
+ )
num_params = count_params(model)
- config['num_params'] = num_params
- print(f'Number of parameters: {num_params}')
+ config["num_params"] = num_params
+ print(f"Number of parameters: {num_params}")
# Load from checkpoint
if args.ckpt:
ckpt_path = args.ckpt
ckpt = paddle.load(ckpt_path)
- model.load_state_dict(ckpt['model'])
- print('Weights loaded from %s' % ckpt_path)
-
+ model.load_state_dict(ckpt["model"])
+ print("Weights loaded from %s" % ckpt_path)
+
if args.test:
- batchsize = config['test']['batchsize']
- testset = KFDataset(paths=config['data']['paths'],
- raw_res=config['data']['raw_res'],
- data_res=config['test']['data_res'],
- pde_res=config['test']['data_res'],
- n_samples=config['data']['n_test_samples'],
- offset=config['data']['testoffset'],
- t_duration=config['data']['t_duration'])
+ batchsize = config["test"]["batchsize"]
+ testset = KFDataset(
+ paths=config["data"]["paths"],
+ raw_res=config["data"]["raw_res"],
+ data_res=config["test"]["data_res"],
+ pde_res=config["test"]["data_res"],
+ n_samples=config["data"]["n_test_samples"],
+ offset=config["data"]["testoffset"],
+ t_duration=config["data"]["t_duration"],
+ )
testloader = DataLoader(testset, batch_size=batchsize, num_workers=4)
criterion = LpLoss()
test_err, std_err = eval_ns(model, testloader, criterion)
- print(f'Averaged test relative L2 error: {test_err}; Standard error: {std_err}')
+ print(f"Averaged test relative L2 error: {test_err}; Standard error: {std_err}")
else:
# training set
- batchsize = config['train']['batchsize']
- u_set = KFDataset(paths=config['data']['paths'],
- raw_res=config['data']['raw_res'],
- data_res=config['data']['data_res'],
- pde_res=config['data']['data_res'],
- n_samples=config['data']['n_data_samples'],
- offset=config['data']['offset'],
- t_duration=config['data']['t_duration'])
+ batchsize = config["train"]["batchsize"]
+ u_set = KFDataset(
+ paths=config["data"]["paths"],
+ raw_res=config["data"]["raw_res"],
+ data_res=config["data"]["data_res"],
+ pde_res=config["data"]["data_res"],
+ n_samples=config["data"]["n_data_samples"],
+ offset=config["data"]["offset"],
+ t_duration=config["data"]["t_duration"],
+ )
u_loader = DataLoader(u_set, batch_size=batchsize, num_workers=4, shuffle=True)
- a_set = KFaDataset(paths=config['data']['paths'],
- raw_res=config['data']['raw_res'],
- pde_res=config['data']['pde_res'],
- n_samples=config['data']['n_a_samples'],
- offset=config['data']['a_offset'],
- t_duration=config['data']['t_duration'])
+ a_set = KFaDataset(
+ paths=config["data"]["paths"],
+ raw_res=config["data"]["raw_res"],
+ pde_res=config["data"]["pde_res"],
+ n_samples=config["data"]["n_a_samples"],
+ offset=config["data"]["a_offset"],
+ t_duration=config["data"]["t_duration"],
+ )
a_loader = DataLoader(a_set, batch_size=batchsize, num_workers=4, shuffle=True)
# val set
- valset = KFDataset(paths=config['data']['paths'],
- raw_res=config['data']['raw_res'],
- data_res=config['test']['data_res'],
- pde_res=config['test']['data_res'],
- n_samples=config['data']['n_test_samples'],
- offset=config['data']['testoffset'],
- t_duration=config['data']['t_duration'])
+ valset = KFDataset(
+ paths=config["data"]["paths"],
+ raw_res=config["data"]["raw_res"],
+ data_res=config["test"]["data_res"],
+ pde_res=config["test"]["data_res"],
+ n_samples=config["data"]["n_test_samples"],
+ offset=config["data"]["testoffset"],
+ t_duration=config["data"]["t_duration"],
+ )
val_loader = DataLoader(valset, batch_size=batchsize, num_workers=4)
- print(f'Train set: {len(u_set)}; Test set: {len(valset)}; IC set: {len(a_set)}')
+ print(f"Train set: {len(u_set)}; Test set: {len(valset)}; IC set: {len(a_set)}")
- scheduler = MultiStepDecay(learning_rate=config['train']['base_lr'],
- milestones=config['train']['milestones'],
- gamma=config['train']['scheduler_gamma'])
+ scheduler = MultiStepDecay(
+ learning_rate=config["train"]["base_lr"],
+ milestones=config["train"]["milestones"],
+ gamma=config["train"]["scheduler_gamma"],
+ )
optimizer = Adam(learning_rate=scheduler, parameters=model.parameters())
if args.ckpt:
ckpt = paddle.load(ckpt_path)
- optimizer.load_state_dict(ckpt['optim'])
- scheduler.load_state_dict(ckpt['scheduler'])
- config['train']['start_iter'] = scheduler.last_epoch
- train_ns(model,
- u_loader, a_loader,
- val_loader,
- optimizer, scheduler,
- config, args)
- print('Done!')
-
-
-
-if __name__ == '__main__':
+ optimizer.load_state_dict(ckpt["optim"])
+ scheduler.load_state_dict(ckpt["scheduler"])
+ config["train"]["start_iter"] = scheduler.last_epoch
+ train_ns(
+ model, u_loader, a_loader, val_loader, optimizer, scheduler, config, args
+ )
+ print("Done!")
+
+
+if __name__ == "__main__":
paddle.backends.cudnn.benchmark = True
# parse options
- parser = ArgumentParser(description='Basic paser')
- parser.add_argument('--config', type=str, help='Path to the configuration file')
- parser.add_argument('--log', action='store_true', help='Turn on the wandb')
- parser.add_argument('--seed', type=int, default=None)
- parser.add_argument('--ckpt', type=str, default=None)
- parser.add_argument('--test', action='store_true', help='Test')
- parser.add_argument('--tqdm', action='store_true', help='Turn on the tqdm')
+ parser = ArgumentParser(description="Basic paser")
+ parser.add_argument("--config", type=str, help="Path to the configuration file")
+ parser.add_argument("--log", action="store_true", help="Turn on the wandb")
+ parser.add_argument("--seed", type=int, default=None)
+ parser.add_argument("--ckpt", type=str, default=None)
+ parser.add_argument("--test", action="store_true", help="Test")
+ parser.add_argument("--tqdm", action="store_true", help="Turn on the tqdm")
args = parser.parse_args()
if args.seed is None:
args.seed = random.randint(0, 100000)
- subprocess(args)
\ No newline at end of file
+ subprocess(args)
diff --git a/jointContribution/PINO/PINO_paddle/train_utils/__init__.py b/jointContribution/PINO/PINO_paddle/train_utils/__init__.py
index 6bcd02cdfa..86b2918b2b 100644
--- a/jointContribution/PINO/PINO_paddle/train_utils/__init__.py
+++ b/jointContribution/PINO/PINO_paddle/train_utils/__init__.py
@@ -1,2 +1,4 @@
-from .datasets import NSLoader, DarcyFlow
-from .losses import get_forcing, LpLoss
\ No newline at end of file
+from .datasets import DarcyFlow
+from .datasets import NSLoader
+from .losses import LpLoss
+from .losses import get_forcing
diff --git a/jointContribution/PINO/PINO_paddle/train_utils/datasets.py b/jointContribution/PINO/PINO_paddle/train_utils/datasets.py
index e95988b684..dd5faad422 100644
--- a/jointContribution/PINO/PINO_paddle/train_utils/datasets.py
+++ b/jointContribution/PINO/PINO_paddle/train_utils/datasets.py
@@ -1,29 +1,34 @@
-import scipy.io
import numpy as np
+import scipy.io
try:
from pyDOE import lhs
+
# Only needed for PINN's dataset
except ImportError:
lhs = None
import paddle
from paddle.io import Dataset
-from .utils import get_grid3d, convert_ic, paddle2dgrid
+
+from .utils import convert_ic
+from .utils import get_grid3d
+from .utils import paddle2dgrid
+
def online_loader(sampler, S, T, time_scale, batchsize=1):
while True:
u0 = sampler.sample(batchsize)
- a = convert_ic(u0, batchsize,
- S, T,
- time_scale=time_scale)
+ a = convert_ic(u0, batchsize, S, T, time_scale=time_scale)
yield a
+
def sample_data(loader):
while True:
for batch in loader:
yield batch
+
class MatReader(object):
def __init__(self, file_path, to_paddle=True, to_cuda=False, to_float=True):
super(MatReader, self).__init__()
@@ -73,8 +78,9 @@ def set_paddle(self, to_paddle):
def set_float(self, to_float):
self.to_float = to_float
+
class BurgersLoader(object):
- def __init__(self, datapath, nx=2 ** 10, nt=100, sub=8, sub_t=1, new=False):
+ def __init__(self, datapath, nx=2**10, nt=100, sub=8, sub_t=1, new=False):
dataloader = MatReader(datapath)
self.sub = sub
self.sub_t = sub_t
@@ -83,24 +89,29 @@ def __init__(self, datapath, nx=2 ** 10, nt=100, sub=8, sub_t=1, new=False):
self.new = new
if new:
self.T += 1
- self.x_data = dataloader.read_field('input')[:, ::sub]
- self.y_data = dataloader.read_field('output')[:, ::sub_t, ::sub]
- self.v = dataloader.read_field('visc').item()
+ self.x_data = dataloader.read_field("input")[:, ::sub]
+ self.y_data = dataloader.read_field("output")[:, ::sub_t, ::sub]
+ self.v = dataloader.read_field("visc").item()
def make_loader(self, n_sample, batch_size, start=0, train=True):
- Xs = self.x_data[start:start + n_sample]
- ys = self.y_data[start:start + n_sample]
+ Xs = self.x_data[start : start + n_sample]
+ ys = self.y_data[start : start + n_sample]
if self.new:
- gridx = paddle.to_tensor(np.linspace(0, 1, self.s + 1)[:-1], dtype='float32')
- gridt = paddle.to_tensor(np.linspace(0, 1, self.T), dtype='float32')
+ gridx = paddle.to_tensor(
+ np.linspace(0, 1, self.s + 1)[:-1], dtype="float32"
+ )
+ gridt = paddle.to_tensor(np.linspace(0, 1, self.T), dtype="float32")
else:
- gridx = paddle.to_tensor(np.linspace(0, 1, self.s), dtype='float32')
- gridt = paddle.to_tensor(np.linspace(0, 1, self.T + 1)[1:], dtype='float32')
+ gridx = paddle.to_tensor(np.linspace(0, 1, self.s), dtype="float32")
+ gridt = paddle.to_tensor(np.linspace(0, 1, self.T + 1)[1:], dtype="float32")
gridx = gridx.reshape([1, 1, self.s])
gridt = gridt.reshape([1, self.T, 1])
Xs = Xs.reshape([n_sample, 1, self.s]).tile([1, self.T, 1])
- Xs = paddle.stack([Xs, gridx.tile([n_sample, self.T, 1]), gridt.tile([n_sample, 1, self.s])], axis=3)
+ Xs = paddle.stack(
+ [Xs, gridx.tile([n_sample, self.T, 1]), gridt.tile([n_sample, 1, self.s])],
+ axis=3,
+ )
dataset = paddle.io.TensorDataset([Xs, ys])
if train:
loader = paddle.io.DataLoader(dataset, batch_size=batch_size, shuffle=False)
@@ -108,12 +119,12 @@ def make_loader(self, n_sample, batch_size, start=0, train=True):
loader = paddle.io.DataLoader(dataset, batch_size=batch_size, shuffle=False)
return loader
+
class NSLoader(object):
- def __init__(self, datapath1,
- nx, nt,
- datapath2=None, sub=1, sub_t=1,
- N=100, t_interval=1.0):
- '''
+ def __init__(
+ self, datapath1, nx, nt, datapath2=None, sub=1, sub_t=1, N=100, t_interval=1.0
+ ):
+ """
Load data from npy and reshape to (N, X, Y, T)
Args:
datapath1: path to data
@@ -124,16 +135,16 @@ def __init__(self, datapath1,
sub_t:
N:
t_interval:
- '''
+ """
self.S = nx // sub
self.T = int(nt * t_interval) // sub_t + 1
self.time_scale = t_interval
data1 = np.load(datapath1)
- data1 = paddle.to_tensor(data1, dtype='float32')[..., ::sub_t, ::sub, ::sub]
+ data1 = paddle.to_tensor(data1, dtype="float32")[..., ::sub_t, ::sub, ::sub]
if datapath2 is not None:
data2 = np.load(datapath2)
- data2 = paddle.to_tensor(data2, dtype='float32')[..., ::sub_t, ::sub, ::sub]
+ data2 = paddle.to_tensor(data2, dtype="float32")[..., ::sub_t, ::sub, ::sub]
if t_interval == 0.5:
data1 = self.extract(data1)
if datapath2 is not None:
@@ -147,46 +158,69 @@ def __init__(self, datapath1,
def make_loader(self, n_sample, batch_size, start=0, train=True):
if train:
- a_data = self.data[start:start + n_sample, :, :, 0].reshape([n_sample, self.S, self.S])
- u_data = self.data[start:start + n_sample].reshape([n_sample, self.S, self.S, self.T])
+ a_data = self.data[start : start + n_sample, :, :, 0].reshape(
+ [n_sample, self.S, self.S]
+ )
+ u_data = self.data[start : start + n_sample].reshape(
+ [n_sample, self.S, self.S, self.T]
+ )
else:
a_data = self.data[-n_sample:, :, :, 0].reshape([n_sample, self.S, self.S])
u_data = self.data[-n_sample:].reshape([n_sample, self.S, self.S, self.T])
- a_data = a_data.reshape([n_sample, self.S, self.S, 1, 1]).tile([1, 1, 1, self.T, 1])
+ a_data = a_data.reshape([n_sample, self.S, self.S, 1, 1]).tile(
+ [1, 1, 1, self.T, 1]
+ )
gridx, gridy, gridt = get_grid3d(self.S, self.T, time_scale=self.time_scale)
- a_data = paddle.concat((gridx.tile([n_sample, 1, 1, 1, 1]), gridy.tile([n_sample, 1, 1, 1, 1]),
- gridt.tile([n_sample, 1, 1, 1, 1]), a_data), axis=-1)
+ a_data = paddle.concat(
+ (
+ gridx.tile([n_sample, 1, 1, 1, 1]),
+ gridy.tile([n_sample, 1, 1, 1, 1]),
+ gridt.tile([n_sample, 1, 1, 1, 1]),
+ a_data,
+ ),
+ axis=-1,
+ )
dataset = paddle.io.TensorDataset(a_data, u_data)
loader = paddle.io.DataLoader(dataset, batch_size=batch_size, shuffle=train)
return loader
def make_dataset(self, n_sample, start=0, train=True):
if train:
- a_data = self.data[start:start + n_sample, :, :, 0].reshape([n_sample, self.S, self.S])
- u_data = self.data[start:start + n_sample].reshape([n_sample, self.S, self.S, self.T])
+ a_data = self.data[start : start + n_sample, :, :, 0].reshape(
+ [n_sample, self.S, self.S]
+ )
+ u_data = self.data[start : start + n_sample].reshape(
+ [n_sample, self.S, self.S, self.T]
+ )
else:
a_data = self.data[-n_sample:, :, :, 0].reshape([n_sample, self.S, self.S])
u_data = self.data[-n_sample:].reshape([n_sample, self.S, self.S, self.T])
- a_data = a_data.reshape([n_sample, self.S, self.S, 1, 1]).tile([1, 1, 1, self.T, 1])
+ a_data = a_data.reshape([n_sample, self.S, self.S, 1, 1]).tile(
+ [1, 1, 1, self.T, 1]
+ )
gridx, gridy, gridt = get_grid3d(self.S, self.T)
- a_data = paddle.concat((
- gridx.tile([n_sample, 1, 1, 1, 1]),
- gridy.tile([n_sample, 1, 1, 1, 1]),
- gridt.tile([n_sample, 1, 1, 1, 1]),
- a_data), axis=-1)
+ a_data = paddle.concat(
+ (
+ gridx.tile([n_sample, 1, 1, 1, 1]),
+ gridy.tile([n_sample, 1, 1, 1, 1]),
+ gridt.tile([n_sample, 1, 1, 1, 1]),
+ a_data,
+ ),
+ axis=-1,
+ )
dataset = paddle.io.TensorDataset(a_data, u_data)
return dataset
@staticmethod
def extract(data):
- '''
+ """
Extract data with time range 0-0.5, 0.25-0.75, 0.5-1.0, 0.75-1.25,...
Args:
data: tensor with size N x 129 x 128 x 128
Returns:
output: (4*N-1) x 65 x 128 x 128
- '''
+ """
T = data.shape[1] // 2
interval = data.shape[1] // 4
N = data.shape[0]
@@ -197,25 +231,34 @@ def extract(data):
# reach boundary
break
if j != 3:
- new_data[i * 4 + j] = data[i, interval * j:interval * j + T + 1]
+ new_data[i * 4 + j] = data[i, interval * j : interval * j + T + 1]
else:
- new_data[i * 4 + j, 0: interval] = data[i, interval * j:interval * j + interval]
- new_data[i * 4 + j, interval: T + 1] = data[i + 1, 0:interval + 1]
+ new_data[i * 4 + j, 0:interval] = data[
+ i, interval * j : interval * j + interval
+ ]
+ new_data[i * 4 + j, interval : T + 1] = data[
+ i + 1, 0 : interval + 1
+ ]
return new_data
+
class KFDataset(Dataset):
- def __init__(self, paths,
- data_res, pde_res,
- raw_res,
- n_samples=None,
- total_samples=None,
- idx=0,
- offset=0,
- t_duration=1.0):
+ def __init__(
+ self,
+ paths,
+ data_res,
+ pde_res,
+ raw_res,
+ n_samples=None,
+ total_samples=None,
+ idx=0,
+ offset=0,
+ t_duration=1.0,
+ ):
super().__init__()
- self.data_res = data_res # data resolution
- self.pde_res = pde_res # pde loss resolution
- self.raw_res = raw_res # raw data resolution
+ self.data_res = data_res # data resolution
+ self.pde_res = pde_res # pde loss resolution
+ self.raw_res = raw_res # raw data resolution
self.t_duration = t_duration
self.paths = paths
self.offset = offset
@@ -223,94 +266,111 @@ def __init__(self, paths,
if t_duration == 1.0:
self.T = self.pde_res[2]
else:
- self.T = int(self.pde_res[2] * t_duration) + 1 # number of points in time dimension
+ self.T = (
+ int(self.pde_res[2] * t_duration) + 1
+ ) # number of points in time dimension
self.load()
if total_samples is not None:
- print(f'Load {total_samples} samples starting from {idx}th sample')
- self.data = self.data[idx:idx + total_samples]
- self.a_data = self.a_data[idx:idx + total_samples]
-
+ print(f"Load {total_samples} samples starting from {idx}th sample")
+ self.data = self.data[idx : idx + total_samples]
+ self.a_data = self.a_data[idx : idx + total_samples]
+
self.data_s_step = pde_res[0] // data_res[0]
self.data_t_step = (pde_res[2] - 1) // (data_res[2] - 1)
def load(self):
datapath = self.paths[0]
- raw_data = np.load(datapath, mmap_mode='r')
+ raw_data = np.load(datapath, mmap_mode="r")
# subsample ratio
sub_x = self.raw_res[0] // self.data_res[0]
sub_t = (self.raw_res[2] - 1) // (self.data_res[2] - 1)
-
+
a_sub_x = self.raw_res[0] // self.pde_res[0]
# load data
- data = raw_data[self.offset: self.offset + self.n_samples, ::sub_t, ::sub_x, ::sub_x]
+ data = raw_data[
+ self.offset : self.offset + self.n_samples, ::sub_t, ::sub_x, ::sub_x
+ ]
# divide data
- if self.t_duration != 0.:
+ if self.t_duration != 0.0:
end_t = self.raw_res[2] - 1
- K = int(1/self.t_duration)
+ K = int(1 / self.t_duration)
step = end_t // K
data = self.partition(data)
- a_data = raw_data[self.offset: self.offset + self.n_samples, 0:end_t:step, ::a_sub_x, ::a_sub_x]
- a_data = a_data.reshape([self.n_samples * K, 1, self.pde_res[0], self.pde_res[1]]) # 2N x 1 x S x S
+ a_data = raw_data[
+ self.offset : self.offset + self.n_samples,
+ 0:end_t:step,
+ ::a_sub_x,
+ ::a_sub_x,
+ ]
+ a_data = a_data.reshape(
+ [self.n_samples * K, 1, self.pde_res[0], self.pde_res[1]]
+ ) # 2N x 1 x S x S
else:
- a_data = raw_data[self.offset: self.offset + self.n_samples, 0:1, ::a_sub_x, ::a_sub_x]
+ a_data = raw_data[
+ self.offset : self.offset + self.n_samples, 0:1, ::a_sub_x, ::a_sub_x
+ ]
# convert into paddle tensor
- data = paddle.to_tensor(data, dtype='float32')
- a_data = paddle.to_tensor(a_data, dtype='float32').transpose([0, 2, 3, 1])
+ data = paddle.to_tensor(data, dtype="float32")
+ a_data = paddle.to_tensor(a_data, dtype="float32").transpose([0, 2, 3, 1])
self.data = data.transpose([0, 2, 3, 1])
S = self.pde_res[1]
-
- a_data = a_data[:, :, :, :, None] # N x S x S x 1 x 1
+
+ a_data = a_data[:, :, :, :, None] # N x S x S x 1 x 1
gridx, gridy, gridt = get_grid3d(S, self.T)
- self.grid = paddle.concat((gridx[0], gridy[0], gridt[0]), axis=-1) # S x S x T x 3
+ self.grid = paddle.concat(
+ (gridx[0], gridy[0], gridt[0]), axis=-1
+ ) # S x S x T x 3
self.a_data = a_data
def partition(self, data):
- '''
+ """
Args:
data: tensor with size N x T x S x S
Returns:
output: int(1/t_duration) *N x (T//2 + 1) x 128 x 128
- '''
+ """
N, T, S = data.shape[:3]
K = int(1 / self.t_duration)
new_data = np.zeros((K * N, T // K + 1, S, S))
step = T // K
for i in range(N):
for j in range(K):
- new_data[i * K + j] = data[i, j * step: (j+1) * step + 1]
+ new_data[i * K + j] = data[i, j * step : (j + 1) * step + 1]
return new_data
def __getitem__(self, idx):
- a_data = paddle.concat((
- self.grid,
- self.a_data[idx].tile(1, 1, self.T, 1)
- ), axis=-1)
+ a_data = paddle.concat(
+ (self.grid, self.a_data[idx].tile(1, 1, self.T, 1)), axis=-1
+ )
return self.data[idx], a_data
- def __len__(self, ):
+ def __len__(
+ self,
+ ):
return self.data.shape[0]
+
class BurgerData(Dataset):
- '''
- members:
+ """
+ members:
- t, x, Exact: raw data
- - X, T: meshgrid
+ - X, T: meshgrid
- X_star, u_star: flattened (x, t), u array
- lb, ub: lower bound and upper bound vector
- X_u, u: boundary condition data (x, t), u
- '''
+ """
def __init__(self, datapath):
data = scipy.io.loadmat(datapath)
# raw 2D data
- self.t = data['t'].flatten()[:, None] # (100,1)
- self.x = data['x'].flatten()[:, None] # (256, 1)
- self.Exact = np.real(data['usol']).T # (100, 256)
+ self.t = data["t"].flatten()[:, None] # (100,1)
+ self.x = data["x"].flatten()[:, None] # (256, 1)
+ self.Exact = np.real(data["usol"]).T # (100, 256)
# Flattened sequence
self.get_flatten_data()
@@ -344,39 +404,40 @@ def get_boundary_data(self):
self.u = np.vstack([uu1, uu2, uu3])
def sample_xt(self, N=10000):
- '''
+ """
Sample (x, t) pairs within the boundary
Return:
- X_f: (N, 2) array
- '''
+ """
X_f = self.lb + (self.ub - self.lb) * lhs(2, N)
X_f = np.vstack((X_f, self.X_u))
return X_f
def sample_xu(self, N=100):
- '''
+ """
Sample N points from boundary data
- Return:
- - X_u: (N, 2) array
+ Return:
+ - X_u: (N, 2) array
- u: (N, 1) array
- '''
+ """
idx = np.random.choice(self.X_u.shape[0], N, replace=False)
X_u = self.X_u[idx, :]
u = self.u[idx, :]
return X_u, u
+
class DarcyFlow(Dataset):
- def __init__(self,
- datapath,
- nx, sub,
- offset=0,
- num=1):
+ def __init__(self, datapath, nx, sub, offset=0, num=1):
self.S = int(nx // sub) + 1 if sub > 1 else nx
data = scipy.io.loadmat(datapath)
- a = data['coeff']
- u = data['sol']
- self.a = paddle.to_tensor(a[offset: offset + num, ::sub, ::sub], dtype='float32')
- self.u = paddle.to_tensor(u[offset: offset + num, ::sub, ::sub], dtype='float32')
+ a = data["coeff"]
+ u = data["sol"]
+ self.a = paddle.to_tensor(
+ a[offset : offset + num, ::sub, ::sub], dtype="float32"
+ )
+ self.u = paddle.to_tensor(
+ u[offset : offset + num, ::sub, ::sub], dtype="float32"
+ )
self.mesh = paddle2dgrid(self.S, self.S)
def __len__(self):
@@ -386,22 +447,25 @@ def __getitem__(self, item):
fa = self.a[item]
return paddle.concat([fa.unsqueeze(2), self.mesh], axis=2), self.u[item]
+
class DarcyIC(Dataset):
- def __init__(self,
- datapath,
- nx, sub,
- offset=0,
- num=1):
+ def __init__(self, datapath, nx, sub, offset=0, num=1):
self.S = int(nx // sub) + 1 if sub > 1 else nx
data = scipy.io.loadmat(datapath)
- a = data['coeff']
- self.a = paddle.to_tensor(a[offset: offset + num, ::sub, ::sub], dtype='float32')
+ a = data["coeff"]
+ self.a = paddle.to_tensor(
+ a[offset : offset + num, ::sub, ::sub], dtype="float32"
+ )
self.mesh = paddle2dgrid(self.S, self.S)
data = scipy.io.loadmat(datapath)
- a = data['coeff']
- u = data['sol']
- self.a = paddle.to_tensor(a[offset: offset + num, ::sub, ::sub], dtype='float32')
- self.u = paddle.to_tensor(u[offset: offset + num, ::sub, ::sub], dtype='float32')
+ a = data["coeff"]
+ u = data["sol"]
+ self.a = paddle.to_tensor(
+ a[offset : offset + num, ::sub, ::sub], dtype="float32"
+ )
+ self.u = paddle.to_tensor(
+ u[offset : offset + num, ::sub, ::sub], dtype="float32"
+ )
self.mesh = paddle2dgrid(self.S, self.S)
def __len__(self):
@@ -409,24 +473,27 @@ def __len__(self):
def __getitem__(self, item):
fa = self.a[item]
- return paddle.concat([fa.unsqueeze(2), self.mesh], axis=2)
+ return paddle.concat([fa.unsqueeze(2), self.mesh], axis=2)
+
class DarcyCombo(Dataset):
- def __init__(self,
- datapath,
- nx,
- sub, pde_sub,
- num=1000, offset=0) -> None:
+ def __init__(self, datapath, nx, sub, pde_sub, num=1000, offset=0) -> None:
super().__init__()
self.S = int(nx // sub) + 1 if sub > 1 else nx
self.pde_S = int(nx // pde_sub) + 1 if sub > 1 else nx
data = scipy.io.loadmat(datapath)
- a = data['coeff']
- u = data['sol']
- self.a = paddle.to_tensor(a[offset: offset + num, ::sub, ::sub], dtype='float32')
- self.u = paddle.to_tensor(u[offset: offset + num, ::sub, ::sub], dtype='float32')
+ a = data["coeff"]
+ u = data["sol"]
+ self.a = paddle.to_tensor(
+ a[offset : offset + num, ::sub, ::sub], dtype="float32"
+ )
+ self.u = paddle.to_tensor(
+ u[offset : offset + num, ::sub, ::sub], dtype="float32"
+ )
self.mesh = paddle2dgrid(self.S, self.S)
- self.pde_a = paddle.to_tensor(a[offset: offset + num, ::pde_sub, ::pde_sub], dtype='float32')
+ self.pde_a = paddle.to_tensor(
+ a[offset : offset + num, ::pde_sub, ::pde_sub], dtype="float32"
+ )
self.pde_mesh = paddle2dgrid(self.pde_S, self.pde_S)
def __len__(self):
@@ -439,19 +506,19 @@ def __getitem__(self, item):
pde_ic = paddle.concat([pde_a.unsqueeze[2], self.pde_mesh], axis=2)
return data_ic, self.u[item], pde_ic
-'''
+
+"""
dataset class for loading initial conditions for Komogrov flow
-'''
+"""
+
+
class KFaDataset(Dataset):
- def __init__(self, paths,
- pde_res,
- raw_res,
- n_samples=None,
- offset=0,
- t_duration=1.0):
+ def __init__(
+ self, paths, pde_res, raw_res, n_samples=None, offset=0, t_duration=1.0
+ ):
super().__init__()
- self.pde_res = pde_res # pde loss resolution
- self.raw_res = raw_res # raw data resolution
+ self.pde_res = pde_res # pde loss resolution
+ self.raw_res = raw_res # raw data resolution
self.t_duration = t_duration
self.paths = paths
self.offset = offset
@@ -459,39 +526,53 @@ def __init__(self, paths,
if t_duration == 1.0:
self.T = self.pde_res[2]
else:
- self.T = int(self.pde_res[2] * t_duration) + 1 # number of points in time dimension
+ self.T = (
+ int(self.pde_res[2] * t_duration) + 1
+ ) # number of points in time dimension
self.load()
def load(self):
datapath = self.paths[0]
- raw_data = np.load(datapath, mmap_mode='r')
+ raw_data = np.load(datapath, mmap_mode="r")
# subsample ratio
a_sub_x = self.raw_res[0] // self.pde_res[0]
# load data
- if self.t_duration != 0.:
+ if self.t_duration != 0.0:
end_t = self.raw_res[2] - 1
- K = int(1/self.t_duration)
+ K = int(1 / self.t_duration)
step = end_t // K
- a_data = raw_data[self.offset: self.offset + self.n_samples, 0:end_t:step, ::a_sub_x, ::a_sub_x]
- a_data = a_data.reshape([self.n_samples * K, 1, self.pde_res[0], self.pde_res[1]]) # 2N x 1 x S x S
+ a_data = raw_data[
+ self.offset : self.offset + self.n_samples,
+ 0:end_t:step,
+ ::a_sub_x,
+ ::a_sub_x,
+ ]
+ a_data = a_data.reshape(
+ [self.n_samples * K, 1, self.pde_res[0], self.pde_res[1]]
+ ) # 2N x 1 x S x S
else:
- a_data = raw_data[self.offset: self.offset + self.n_samples, 0:1, ::a_sub_x, ::a_sub_x]
+ a_data = raw_data[
+ self.offset : self.offset + self.n_samples, 0:1, ::a_sub_x, ::a_sub_x
+ ]
# convert into tensor
- a_data = paddle.to_tensor(a_data, dtype='float32').permute(0, 2, 3, 1)
+ a_data = paddle.to_tensor(a_data, dtype="float32").permute(0, 2, 3, 1)
S = self.pde_res[1]
- a_data = a_data[:, :, :, :, None] # N x S x S x 1 x 1
+ a_data = a_data[:, :, :, :, None] # N x S x S x 1 x 1
gridx, gridy, gridt = get_grid3d(S, self.T)
- self.grid = paddle.concat((gridx[0], gridy[0], gridt[0]), axis=-1) # S x S x T x 3
+ self.grid = paddle.concat(
+ (gridx[0], gridy[0], gridt[0]), axis=-1
+ ) # S x S x T x 3
self.a_data = a_data
def __getitem__(self, idx):
- a_data = paddle.concat((
- self.grid,
- self.a_data[idx].tile(1, 1, self.T, 1)
- ), axis=-1)
+ a_data = paddle.concat(
+ (self.grid, self.a_data[idx].tile(1, 1, self.T, 1)), axis=-1
+ )
return a_data
- def __len__(self, ):
- return self.a_data.shape[0]
\ No newline at end of file
+ def __len__(
+ self,
+ ):
+ return self.a_data.shape[0]
diff --git a/jointContribution/PINO/PINO_paddle/train_utils/eval_2d.py b/jointContribution/PINO/PINO_paddle/train_utils/eval_2d.py
index 459c97d3a9..b5e0e08a4a 100644
--- a/jointContribution/PINO/PINO_paddle/train_utils/eval_2d.py
+++ b/jointContribution/PINO/PINO_paddle/train_utils/eval_2d.py
@@ -1,16 +1,14 @@
-from tqdm import tqdm
+import matplotlib.pyplot as plt
import numpy as np
-
import paddle
+from tqdm import tqdm
-from .losses import LpLoss, darcy_loss, PINO_loss
+from .losses import LpLoss
+from .losses import PINO_loss
+from .losses import darcy_loss
-import matplotlib.pyplot as plt
-def eval_darcy(model,
- dataloader,
- config,
- use_tqdm=True):
+def eval_darcy(model, dataloader, config, use_tqdm=True):
model.eval()
myloss = LpLoss(size_average=True)
if use_tqdm:
@@ -19,11 +17,13 @@ def eval_darcy(model,
pbar = dataloader
mesh = dataloader.dataset.mesh
- mollifier = paddle.sin(np.pi * mesh[..., 0]) * paddle.sin(np.pi * mesh[..., 1]) * 0.001
+ mollifier = (
+ paddle.sin(np.pi * mesh[..., 0]) * paddle.sin(np.pi * mesh[..., 1]) * 0.001
+ )
f_val = []
test_err = []
i = 0
- fig, ax = plt.subplots(3,3)
+ fig, ax = plt.subplots(3, 3)
with paddle.no_grad():
for x, y in pbar:
@@ -31,19 +31,19 @@ def eval_darcy(model,
pred = pred * mollifier
if i < 3:
ax[2][i].imshow(pred[0, :, :])
- ax[2][i].set_title('prediction')
+ ax[2][i].set_title("prediction")
ax[1][i].imshow(y[0, :, :])
- ax[1][i].set_title('ground truth')
+ ax[1][i].set_title("ground truth")
ax[0][i].imshow(x[0, :, :, 0])
- ax[0][i].set_title('input')
+ ax[0][i].set_title("input")
for k in range(3):
- ax[k][i].set_xlabel('x')
- ax[k][i].set_ylabel('y')
- if i==3:
+ ax[k][i].set_xlabel("x")
+ ax[k][i].set_ylabel("y")
+ if i == 3:
plt.tight_layout()
- plt.savefig('result.png')
- i+=1
+ plt.savefig("result.png")
+ i += 1
data_loss = myloss(pred, y)
a = x[..., 0]
f_loss = darcy_loss(pred, a)
@@ -53,7 +53,7 @@ def eval_darcy(model,
if use_tqdm:
pbar.set_description(
(
- f'Equation error: {f_loss.item():.5f}, test l2 error: {data_loss.item()}'
+ f"Equation error: {f_loss.item():.5f}, test l2 error: {data_loss.item()}"
)
)
mean_f_err = np.mean(f_val)
@@ -62,14 +62,13 @@ def eval_darcy(model,
mean_err = np.mean(test_err)
std_err = np.std(test_err, ddof=1) / np.sqrt(len(test_err))
- print(f'==Averaged relative L2 error mean: {mean_err}, std error: {std_err}==\n'
- f'==Averaged equation error mean: {mean_f_err}, std error: {std_f_err}==')
+ print(
+ f"==Averaged relative L2 error mean: {mean_err}, std error: {std_err}==\n"
+ f"==Averaged equation error mean: {mean_f_err}, std error: {std_f_err}=="
+ )
+
-def eval_burgers(model,
- dataloader,
- v,
- config,
- use_tqdm=True):
+def eval_burgers(model, dataloader, v, config, use_tqdm=True):
model.eval()
myloss = LpLoss(size_average=True)
if use_tqdm:
@@ -80,24 +79,24 @@ def eval_burgers(model,
test_err = []
f_err = []
i = 0
- fig, ax = plt.subplots(2,3)
+ fig, ax = plt.subplots(2, 3)
for x, y in pbar:
x, y = x, y
out = model(x).reshape(y.shape)
data_loss = myloss(out, y)
- if i<3:
+ if i < 3:
ax[0][i].imshow(out[0, :, :])
- ax[0][i].set_xlabel('x')
- ax[0][i].set_ylabel('t')
- ax[0][i].set_title('prediction')
+ ax[0][i].set_xlabel("x")
+ ax[0][i].set_ylabel("t")
+ ax[0][i].set_title("prediction")
ax[1][i].imshow(y[0, :, :])
- ax[1][i].set_xlabel('x')
- ax[1][i].set_ylabel('t')
- ax[1][i].set_title('ground truth')
- if i==3:
+ ax[1][i].set_xlabel("x")
+ ax[1][i].set_ylabel("t")
+ ax[1][i].set_title("ground truth")
+ if i == 3:
plt.tight_layout()
- plt.savefig('result.png')
- i+=1
+ plt.savefig("result.png")
+ i += 1
loss_u, f_loss = PINO_loss(out, x[:, 0, :, 0], v)
test_err.append(data_loss.item())
f_err.append(f_loss.item())
@@ -108,6 +107,7 @@ def eval_burgers(model,
mean_err = np.mean(test_err)
std_err = np.std(test_err, ddof=1) / np.sqrt(len(test_err))
- print(f'==Averaged relative L2 error mean: {mean_err}, std error: {std_err}==\n'
- f'==Averaged equation error mean: {mean_f_err}, std error: {std_f_err}==')
-
+ print(
+ f"==Averaged relative L2 error mean: {mean_err}, std error: {std_err}==\n"
+ f"==Averaged equation error mean: {mean_f_err}, std error: {std_f_err}=="
+ )
diff --git a/jointContribution/PINO/PINO_paddle/train_utils/eval_3d.py b/jointContribution/PINO/PINO_paddle/train_utils/eval_3d.py
index f387669d1e..5f7ff56e46 100644
--- a/jointContribution/PINO/PINO_paddle/train_utils/eval_3d.py
+++ b/jointContribution/PINO/PINO_paddle/train_utils/eval_3d.py
@@ -1,31 +1,35 @@
+from timeit import default_timer
+
import paddle
import paddle.nn.functional as F
-
from tqdm import tqdm
-from timeit import default_timer
-from .losses import LpLoss, PINO_loss3d
+from .losses import LpLoss
+from .losses import PINO_loss3d
+
-def eval_ns(model,
- loader,
- dataloader,
- forcing,
- config,
- device,
- log=False,
- project='PINO-default',
- group='FDM',
- tags=['Nan'],
- use_tqdm=True):
- '''
+def eval_ns(
+ model,
+ loader,
+ dataloader,
+ forcing,
+ config,
+ device,
+ log=False,
+ project="PINO-default",
+ group="FDM",
+ tags=["Nan"],
+ use_tqdm=True,
+):
+ """
Evaluate the model for Navier Stokes equation
- '''
+ """
# data parameters
- v = 1 / config['data']['Re']
+ v = 1 / config["data"]["Re"]
S, T = loader.S, loader.T
- t_interval = config['data']['time_interval']
+ t_interval = config["data"]["time_interval"]
# eval settings
- batch_size = config['test']['batchsize']
+ batch_size = config["test"]["batchsize"]
model.eval()
myloss = LpLoss(size_average=True)
@@ -33,8 +37,7 @@ def eval_ns(model,
pbar = tqdm(dataloader, dynamic_ncols=True, smoothing=0.05)
else:
pbar = dataloader
- loss_dict = {'f_error': 0.0,
- 'test_l2': 0.0}
+ loss_dict = {"f_error": 0.0, "test_l2": 0.0}
start_time = default_timer()
with paddle.no_grad():
for x, y in pbar:
@@ -44,19 +47,23 @@ def eval_ns(model,
out = out[..., :-5]
x = x[:, :, :, 0, -1]
loss_l2 = myloss(out.view(batch_size, S, S, T), y.view(batch_size, S, S, T))
- loss_ic, loss_f = PINO_loss3d(out.view(batch_size, S, S, T), x, forcing, v, t_interval)
+ loss_ic, loss_f = PINO_loss3d(
+ out.view(batch_size, S, S, T), x, forcing, v, t_interval
+ )
- loss_dict['f_error'] += loss_f
- loss_dict['test_l2'] += loss_l2
+ loss_dict["f_error"] += loss_f
+ loss_dict["test_l2"] += loss_l2
if device == 0 and use_tqdm:
pbar.set_description(
(
- f'Train f error: {loss_f.item():.5f}; Test l2 error: {loss_l2.item():.5f}'
+ f"Train f error: {loss_f.item():.5f}; Test l2 error: {loss_l2.item():.5f}"
)
)
end_time = default_timer()
- test_l2 = loss_dict['test_l2'].item() / len(dataloader)
- loss_f = loss_dict['f_error'].item() / len(dataloader)
- print(f'==Averaged relative L2 error is: {test_l2}==\n'
- f'==Averaged equation error is: {loss_f}==')
- print(f'Time cost: {end_time - start_time} s')
+ test_l2 = loss_dict["test_l2"].item() / len(dataloader)
+ loss_f = loss_dict["f_error"].item() / len(dataloader)
+ print(
+ f"==Averaged relative L2 error is: {test_l2}==\n"
+ f"==Averaged equation error is: {loss_f}=="
+ )
+ print(f"Time cost: {end_time - start_time} s")
diff --git a/jointContribution/PINO/PINO_paddle/train_utils/losses.py b/jointContribution/PINO/PINO_paddle/train_utils/losses.py
index 86bc2844f2..ede76a3b88 100644
--- a/jointContribution/PINO/PINO_paddle/train_utils/losses.py
+++ b/jointContribution/PINO/PINO_paddle/train_utils/losses.py
@@ -2,6 +2,7 @@
import paddle
import paddle.nn.functional as F
+
def FDM_Darcy(u, a, D=1):
batchsize = u.size(0)
size = u.size(1)
@@ -18,9 +19,10 @@ def FDM_Darcy(u, a, D=1):
auy = a * uy
auxx = (aux[:, 2:, 1:-1] - aux[:, :-2, 1:-1]) / (2 * dx)
auyy = (auy[:, 1:-1, 2:] - auy[:, 1:-1, :-2]) / (2 * dy)
- Du = - (auxx + auyy)
+ Du = -(auxx + auyy)
return Du
+
def darcy_loss(u, a):
batchsize = u.shape[0]
size = u.shape[1]
@@ -33,7 +35,8 @@ def darcy_loss(u, a):
loss_f = lploss.rel(Du, f)
return loss_f
-def FDM_NS_vorticity(w, v=1/40, t_interval=1.0):
+
+def FDM_NS_vorticity(w, v=1 / 40, t_interval=1.0):
batchsize = w.shape[0]
nx = w.shape[1]
ny = w.shape[2]
@@ -42,14 +45,34 @@ def FDM_NS_vorticity(w, v=1/40, t_interval=1.0):
w_h = paddle.fft.fft2(w, axes=[1, 2])
# Wavenumbers in y-direction
- k_max = nx//2
+ k_max = nx // 2
N = nx
- k_x = paddle.concat((paddle.arange(start=0, end=k_max, step=1),
- paddle.arange(start=-k_max, end=0, step=1)), 0).reshape([N, 1]).tile([1, N]).reshape([1,N,N,1])
- k_y = paddle.concat((paddle.arange(start=0, end=k_max, step=1),
- paddle.arange(start=-k_max, end=0, step=1)), 0).reshape([1, N]).tile([N, 1]).reshape([1,N,N,1])
+ k_x = (
+ paddle.concat(
+ (
+ paddle.arange(start=0, end=k_max, step=1),
+ paddle.arange(start=-k_max, end=0, step=1),
+ ),
+ 0,
+ )
+ .reshape([N, 1])
+ .tile([1, N])
+ .reshape([1, N, N, 1])
+ )
+ k_y = (
+ paddle.concat(
+ (
+ paddle.arange(start=0, end=k_max, step=1),
+ paddle.arange(start=-k_max, end=0, step=1),
+ ),
+ 0,
+ )
+ .reshape([1, N])
+ .tile([N, 1])
+ .reshape([1, N, N, 1])
+ )
# Negative Laplacian in Fourier space
- lap = (k_x ** 2 + k_y ** 2)
+ lap = k_x**2 + k_y**2
lap[0, 0, 0, 0] = 1.0
f_h = w_h / lap
@@ -59,28 +82,31 @@ def FDM_NS_vorticity(w, v=1/40, t_interval=1.0):
wy_h = 1j * k_y * w_h
wlap_h = -lap * w_h
- ux = paddle.fft.irfft2(ux_h[:, :, :k_max + 1], axes=[1, 2])
- uy = paddle.fft.irfft2(uy_h[:, :, :k_max + 1], axes=[1, 2])
- wx = paddle.fft.irfft2(wx_h[:, :, :k_max+1], axes=[1,2])
- wy = paddle.fft.irfft2(wy_h[:, :, :k_max+1], axes=[1,2])
- wlap = paddle.fft.irfft2(wlap_h[:, :, :k_max+1], axes=[1,2])
+ ux = paddle.fft.irfft2(ux_h[:, :, : k_max + 1], axes=[1, 2])
+ uy = paddle.fft.irfft2(uy_h[:, :, : k_max + 1], axes=[1, 2])
+ wx = paddle.fft.irfft2(wx_h[:, :, : k_max + 1], axes=[1, 2])
+ wy = paddle.fft.irfft2(wy_h[:, :, : k_max + 1], axes=[1, 2])
+ wlap = paddle.fft.irfft2(wlap_h[:, :, : k_max + 1], axes=[1, 2])
- dt = t_interval / (nt-1)
+ dt = t_interval / (nt - 1)
wt = (w[:, :, :, 2:] - w[:, :, :, :-2]) / (2 * dt)
- Du1 = wt + (ux*wx + uy*wy - v*wlap)[...,1:-1] #- forcing
+ Du1 = wt + (ux * wx + uy * wy - v * wlap)[..., 1:-1] # - forcing
return Du1
-def Autograd_Burgers(u, grid, v=1/100):
+
+def Autograd_Burgers(u, grid, v=1 / 100):
from paddle.autograd import grad
+
gridt, gridx = grid
ut = grad(u.sum(), gridt, create_graph=True)[0]
ux = grad(u.sum(), gridx, create_graph=True)[0]
uxx = grad(ux.sum(), gridx, create_graph=True)[0]
- Du = ut + ux*u - v*uxx
+ Du = ut + ux * u - v * uxx
return Du, ux, uxx, ut
+
def AD_loss(u, u0, grid, index_ic=None, p=None, q=None):
batchsize = u.size(0)
@@ -92,7 +118,9 @@ def AD_loss(u, u0, grid, index_ic=None, p=None, q=None):
nx = u.size(2)
u = u.reshape(batchsize, nt, nx)
- index_t = paddle.zeros(nx,).long()
+ index_t = paddle.zeros(
+ nx,
+ ).long()
index_x = paddle.tensor(range(nx)).long()
boundary_u = u[:, index_t, index_x]
else:
@@ -106,14 +134,16 @@ def AD_loss(u, u0, grid, index_ic=None, p=None, q=None):
loss_f = F.mse_loss(Du, f)
return loss_ic, loss_f
+
class LpLoss(object):
- '''
+ """
loss function with rel/abs Lp loss
- '''
+ """
+
def __init__(self, d=2, p=2, size_average=True, reduction=True):
super(LpLoss, self).__init__()
- #Dimension and Lp-norm type are postive
+ # Dimension and Lp-norm type are postive
assert d > 0 and p > 0
self.d = d
@@ -124,10 +154,12 @@ def __init__(self, d=2, p=2, size_average=True, reduction=True):
def abs(self, x, y):
num_examples = x.size()[0]
- #Assume uniform mesh
+ # Assume uniform mesh
h = 1.0 / (x.shape[1] - 1.0)
- all_norms = (h**(self.d/self.p))*paddle.norm(x.reshape(num_examples,-1) - y.reshape(num_examples,-1), self.p, 1)
+ all_norms = (h ** (self.d / self.p)) * paddle.norm(
+ x.reshape(num_examples, -1) - y.reshape(num_examples, -1), self.p, 1
+ )
if self.reduction:
if self.size_average:
@@ -140,42 +172,51 @@ def abs(self, x, y):
def rel(self, x, y):
num_examples = x.shape[0]
- diff_norms = paddle.norm(x.reshape([num_examples,-1]) - y.reshape([num_examples,-1]), self.p, 1)
- y_norms = paddle.norm(y.reshape([num_examples,-1]), self.p, 1)
+ diff_norms = paddle.norm(
+ x.reshape([num_examples, -1]) - y.reshape([num_examples, -1]), self.p, 1
+ )
+ y_norms = paddle.norm(y.reshape([num_examples, -1]), self.p, 1)
if self.reduction:
if self.size_average:
- return paddle.mean(diff_norms/y_norms)
+ return paddle.mean(diff_norms / y_norms)
else:
- return paddle.sum(diff_norms/y_norms)
+ return paddle.sum(diff_norms / y_norms)
- return diff_norms/y_norms
+ return diff_norms / y_norms
def __call__(self, x, y):
return self.rel(x, y)
+
def FDM_Burgers(u, v, D=1):
batchsize = u.shape[0]
nt = u.shape[1]
nx = u.shape[2]
u = u.reshape([batchsize, nt, nx])
- dt = D / (nt-1)
- dx = D / (nx)
+ dt = D / (nt - 1)
+ D / (nx)
u_h = paddle.fft.fft(u, axis=2)
# Wavenumbers in y-direction
- k_max = nx//2
- k_x = paddle.concat((paddle.arange(start=0, end=k_max, step=1, dtype='float32'),
- paddle.arange(start=-k_max, end=0, step=1, dtype='float32')), 0).reshape([1,1,nx])
- ux_h = 2j *np.pi*k_x*u_h
- uxx_h = 2j *np.pi*k_x*ux_h
- ux = paddle.fft.irfft(ux_h[:, :, :k_max+1], axis=2, n=nx)
- uxx = paddle.fft.irfft(uxx_h[:, :, :k_max+1], axis=2, n=nx)
+ k_max = nx // 2
+ k_x = paddle.concat(
+ (
+ paddle.arange(start=0, end=k_max, step=1, dtype="float32"),
+ paddle.arange(start=-k_max, end=0, step=1, dtype="float32"),
+ ),
+ 0,
+ ).reshape([1, 1, nx])
+ ux_h = 2j * np.pi * k_x * u_h
+ uxx_h = 2j * np.pi * k_x * ux_h
+ ux = paddle.fft.irfft(ux_h[:, :, : k_max + 1], axis=2, n=nx)
+ uxx = paddle.fft.irfft(uxx_h[:, :, : k_max + 1], axis=2, n=nx)
ut = (u[:, 2:, :] - u[:, :-2, :]) / (2 * dt)
- Du = ut + (ux*u - v*uxx)[:,1:-1,:]
+ Du = ut + (ux * u - v * uxx)[:, 1:-1, :]
return Du
+
def PINO_loss(u, u0, v):
batchsize = u.shape[0]
nt = u.shape[1]
@@ -183,8 +224,8 @@ def PINO_loss(u, u0, v):
u = u.reshape([batchsize, nt, nx])
- index_t = paddle.zeros(1,'int32')
- index_x = paddle.to_tensor(list(range(nx)),'int32')
+ index_t = paddle.zeros(1, "int32")
+ paddle.to_tensor(list(range(nx)), "int32")
boundary_u = paddle.index_select(u, index_t, axis=1).squeeze(1)
loss_u = F.mse_loss(boundary_u, u0)
@@ -194,7 +235,8 @@ def PINO_loss(u, u0, v):
return loss_u, loss_f
-def PINO_loss3d(u, u0, forcing, v=1/40, t_interval=1.0):
+
+def PINO_loss3d(u, u0, forcing, v=1 / 40, t_interval=1.0):
batchsize = u.shape[0]
nx = u.shape[1]
ny = u.shape[2]
@@ -207,13 +249,14 @@ def PINO_loss3d(u, u0, forcing, v=1/40, t_interval=1.0):
loss_ic = lploss(u_in, u0)
Du = FDM_NS_vorticity(u, v, t_interval)
- f = forcing.tile([batchsize, 1, 1, nt-2])
+ f = forcing.tile([batchsize, 1, 1, nt - 2])
loss_f = lploss(Du, f)
return loss_ic, loss_f
+
def PDELoss(model, x, t, nu):
- '''
+ """
Compute the residual of PDE:
residual = u_t + u * u_x - nu * u_{xx} : (N,1)
@@ -223,18 +266,35 @@ def PDELoss(model, x, t, nu):
- nu: constant of PDE
Return:
- mean of residual : scalar
- '''
+ """
u = model(paddle.cat([x, t], dim=1))
# First backward to compute u_x (shape: N x 1), u_t (shape: N x 1)
- grad_x, grad_t = paddle.autograd.grad(outputs=[u.sum()], inputs=[x, t], create_graph=True)
+ grad_x, grad_t = paddle.autograd.grad(
+ outputs=[u.sum()], inputs=[x, t], create_graph=True
+ )
# Second backward to compute u_{xx} (shape N x 1)
- gradgrad_x, = paddle.autograd.grad(outputs=[grad_x.sum()], inputs=[x], create_graph=True)
+ (gradgrad_x,) = paddle.autograd.grad(
+ outputs=[grad_x.sum()], inputs=[x], create_graph=True
+ )
residual = grad_t + u * grad_x - nu * gradgrad_x
return residual
+
def get_forcing(S):
- x1 = paddle.to_tensor(np.linspace(0, 2*np.pi, S, endpoint=False), dtype=paddle.float32).reshape([S, 1]).tile([1, S])
- x2 = paddle.to_tensor(np.linspace(0, 2*np.pi, S, endpoint=False), dtype=paddle.float32).reshape([1, S]).tile([S, 1])
- return -4 * (paddle.cos(4*(x2))).reshape([1,S,S,1])
\ No newline at end of file
+ (
+ paddle.to_tensor(
+ np.linspace(0, 2 * np.pi, S, endpoint=False), dtype=paddle.float32
+ )
+ .reshape([S, 1])
+ .tile([1, S])
+ )
+ x2 = (
+ paddle.to_tensor(
+ np.linspace(0, 2 * np.pi, S, endpoint=False), dtype=paddle.float32
+ )
+ .reshape([1, S])
+ .tile([S, 1])
+ )
+ return -4 * (paddle.cos(4 * (x2))).reshape([1, S, S, 1])
diff --git a/jointContribution/PINO/PINO_paddle/train_utils/train_2d.py b/jointContribution/PINO/PINO_paddle/train_utils/train_2d.py
index 3e41fc590a..daaa1feec2 100644
--- a/jointContribution/PINO/PINO_paddle/train_utils/train_2d.py
+++ b/jointContribution/PINO/PINO_paddle/train_utils/train_2d.py
@@ -1,36 +1,52 @@
import numpy as np
import paddle
from tqdm import tqdm
+
+from .losses import LpLoss
+from .losses import PINO_loss
+from .losses import darcy_loss
from .utils import save_checkpoint
-from .losses import LpLoss, darcy_loss, PINO_loss
-
-def train_2d_operator(model,
- train_loader,
- optimizer, scheduler,
- config,
- rank=0, log=False,
- project='PINO-2d-default',
- group='default',
- tags=['default'],
- use_tqdm=True,
- profile=False):
-
- data_weight = config['train']['xy_loss']
- f_weight = config['train']['f_loss']
+
+
+def train_2d_operator(
+ model,
+ train_loader,
+ optimizer,
+ scheduler,
+ config,
+ rank=0,
+ log=False,
+ project="PINO-2d-default",
+ group="default",
+ tags=["default"],
+ use_tqdm=True,
+ profile=False,
+):
+
+ data_weight = config["train"]["xy_loss"]
+ f_weight = config["train"]["f_loss"]
model.train()
myloss = LpLoss(size_average=True)
- pbar = range(config['train']['epochs'])
+ pbar = range(config["train"]["epochs"])
if use_tqdm:
pbar = tqdm(pbar, dynamic_ncols=True, smoothing=0.1)
mesh = train_loader.dataset.mesh
- mollifier = paddle.sin(np.pi * mesh[..., 0]) * paddle.sin(np.pi * mesh[..., 1]) * 0.001
+ mollifier = (
+ paddle.sin(np.pi * mesh[..., 0]) * paddle.sin(np.pi * mesh[..., 1]) * 0.001
+ )
pde_mesh = train_loader.dataset.pde_mesh
- pde_mol = paddle.sin(np.pi * pde_mesh[..., 0]) * paddle.sin(np.pi * pde_mesh[..., 1]) * 0.001
+ (
+ paddle.sin(np.pi * pde_mesh[..., 0])
+ * paddle.sin(np.pi * pde_mesh[..., 1])
+ * 0.001
+ )
for e in pbar:
- loss_dict = {'train_loss': 0.0,
- 'data_loss': 0.0,
- 'f_loss': 0.0,
- 'test_error': 0.0}
+ loss_dict = {
+ "train_loss": 0.0,
+ "data_loss": 0.0,
+ "f_loss": 0.0,
+ "test_error": 0.0,
+ }
for data_ic, u, pde_ic in train_loader:
data_ic, u, pde_ic = data_ic.to(rank), u.to(rank), pde_ic.to(rank)
@@ -49,44 +65,50 @@ def train_2d_operator(model,
loss.backward()
optimizer.step()
- loss_dict['train_loss'] += loss.item() * y.shape[0]
- loss_dict['f_loss'] += f_loss.item() * y.shape[0]
- loss_dict['data_loss'] += data_loss.item() * y.shape[0]
+ loss_dict["train_loss"] += loss.item() * y.shape[0]
+ loss_dict["f_loss"] += f_loss.item() * y.shape[0]
+ loss_dict["data_loss"] += data_loss.item() * y.shape[0]
scheduler.step()
- train_loss_val = loss_dict['train_loss'] / len(train_loader.dataset)
- f_loss_val = loss_dict['f_loss'] / len(train_loader.dataset)
- data_loss_val = loss_dict['data_loss'] / len(train_loader.dataset)
+ train_loss_val = loss_dict["train_loss"] / len(train_loader.dataset)
+ f_loss_val = loss_dict["f_loss"] / len(train_loader.dataset)
+ data_loss_val = loss_dict["data_loss"] / len(train_loader.dataset)
if use_tqdm:
pbar.set_description(
(
- f'Epoch: {e}, train loss: {train_loss_val:.5f}, '
- f'f_loss: {f_loss_val:.5f}, '
- f'data loss: {data_loss_val:.5f}'
+ f"Epoch: {e}, train loss: {train_loss_val:.5f}, "
+ f"f_loss: {f_loss_val:.5f}, "
+ f"data loss: {data_loss_val:.5f}"
)
)
- save_checkpoint(config['train']['save_dir'],
- config['train']['save_name'],
- model, optimizer)
- print('Done!')
-
-def train_2d_burger(model,
- train_loader, v,
- optimizer, scheduler,
- config,
- rank=0, log=False,
- project='PINO-2d-default',
- group='default',
- tags=['default'],
- use_tqdm=True):
-
- data_weight = config['train']['xy_loss']
- f_weight = config['train']['f_loss']
- ic_weight = config['train']['ic_loss']
+ save_checkpoint(
+ config["train"]["save_dir"], config["train"]["save_name"], model, optimizer
+ )
+ print("Done!")
+
+
+def train_2d_burger(
+ model,
+ train_loader,
+ v,
+ optimizer,
+ scheduler,
+ config,
+ rank=0,
+ log=False,
+ project="PINO-2d-default",
+ group="default",
+ tags=["default"],
+ use_tqdm=True,
+):
+
+ data_weight = config["train"]["xy_loss"]
+ f_weight = config["train"]["f_loss"]
+ ic_weight = config["train"]["ic_loss"]
model.train()
myloss = LpLoss(size_average=True)
- pbar = range(config['train']['epochs'])
+ pbar = range(config["train"]["epochs"])
if use_tqdm:
pbar = tqdm(pbar, dynamic_ncols=True, smoothing=0.1)
@@ -102,8 +124,10 @@ def train_2d_burger(model,
data_loss = myloss(out, y)
loss_u, loss_f = PINO_loss(out, x[:, 0, :, 0], v)
- total_loss = loss_u * ic_weight + loss_f * f_weight + data_loss * data_weight
-
+ total_loss = (
+ loss_u * ic_weight + loss_f * f_weight + data_loss * data_weight
+ )
+
optimizer.clear_grad()
total_loss.backward()
optimizer.step()
@@ -111,7 +135,7 @@ def train_2d_burger(model,
data_l2 += data_loss.item()
train_pino += loss_f.item()
train_loss += total_loss.item()
-
+
scheduler.step()
data_l2 /= len(train_loader)
train_pino /= len(train_loader)
@@ -119,17 +143,20 @@ def train_2d_burger(model,
if use_tqdm:
pbar.set_description(
(
- f'Epoch {e}, train loss: {train_loss:.5f} '
- f'train f error: {train_pino:.5f}; '
- f'data l2 error: {data_l2:.5f}'
+ f"Epoch {e}, train loss: {train_loss:.5f} "
+ f"train f error: {train_pino:.5f}; "
+ f"data l2 error: {data_l2:.5f}"
)
)
if e % 100 == 0:
- save_checkpoint(config['train']['save_dir'],
- config['train']['save_name'].replace('.pt', f'_{e}.pt'),
- model, optimizer)
- save_checkpoint(config['train']['save_dir'],
- config['train']['save_name'],
- model, optimizer)
- print('Done!')
\ No newline at end of file
+ save_checkpoint(
+ config["train"]["save_dir"],
+ config["train"]["save_name"].replace(".pt", f"_{e}.pt"),
+ model,
+ optimizer,
+ )
+ save_checkpoint(
+ config["train"]["save_dir"], config["train"]["save_name"], model, optimizer
+ )
+ print("Done!")
diff --git a/jointContribution/PINO/PINO_paddle/train_utils/train_3d.py b/jointContribution/PINO/PINO_paddle/train_utils/train_3d.py
index 87b2c6c485..fe7a201fdb 100644
--- a/jointContribution/PINO/PINO_paddle/train_utils/train_3d.py
+++ b/jointContribution/PINO/PINO_paddle/train_utils/train_3d.py
@@ -1,53 +1,58 @@
-import paddle
-from tqdm import tqdm
from timeit import default_timer
+
+import paddle
import paddle.nn.functional as F
-from .utils import save_checkpoint
-from .losses import LpLoss, PINO_loss3d, get_forcing
-from .distributed import reduce_loss_dict
+from tqdm import tqdm
+
from .data_utils import sample_data
+from .distributed import reduce_loss_dict
+from .losses import LpLoss
+from .losses import PINO_loss3d
+from .losses import get_forcing
+from .utils import save_checkpoint
-
-def train(model,
- loader, train_loader,
- optimizer, scheduler,
- forcing, config,
- rank=0,
- log=False,
- project='PINO-default',
- group='FDM',
- tags=['Nan'],
- use_tqdm=True,
- profile=False):
+def train(
+ model,
+ loader,
+ train_loader,
+ optimizer,
+ scheduler,
+ forcing,
+ config,
+ rank=0,
+ log=False,
+ project="PINO-default",
+ group="FDM",
+ tags=["Nan"],
+ use_tqdm=True,
+ profile=False,
+):
# data parameters
- v = 1 / config['data']['Re']
+ v = 1 / config["data"]["Re"]
S, T = loader.S, loader.T
- t_interval = config['data']['time_interval']
+ t_interval = config["data"]["time_interval"]
# training settings
- batch_size = config['train']['batchsize']
- ic_weight = config['train']['ic_loss']
- f_weight = config['train']['f_loss']
- xy_weight = config['train']['xy_loss']
+ batch_size = config["train"]["batchsize"]
+ ic_weight = config["train"]["ic_loss"]
+ f_weight = config["train"]["f_loss"]
+ xy_weight = config["train"]["xy_loss"]
model.train()
myloss = LpLoss(size_average=True)
- pbar = range(config['train']['epochs'])
+ pbar = range(config["train"]["epochs"])
if use_tqdm:
pbar = tqdm(pbar, dynamic_ncols=True, smoothing=0.05)
- zero = paddle.zeros(1, dtype='float32').to(rank)
+ zero = paddle.zeros(1, dtype="float32").to(rank)
for ep in pbar:
- loss_dict = {'train_loss': 0.0,
- 'train_ic': 0.0,
- 'train_f': 0.0,
- 'test_l2': 0.0}
+ loss_dict = {"train_loss": 0.0, "train_ic": 0.0, "train_f": 0.0, "test_l2": 0.0}
log_dict = {}
if rank == 0 and profile:
- paddle.cuda.synchronize()
- t1 = default_timer()
+ paddle.cuda.synchronize()
+ t1 = default_timer()
# start solving
for x, y in train_loader:
x, y = x.to(rank), y.to(rank)
@@ -61,7 +66,9 @@ def train(model,
loss_l2 = myloss(out.view(batch_size, S, S, T), y.view(batch_size, S, S, T))
if ic_weight != 0 or f_weight != 0:
- loss_ic, loss_f = PINO_loss3d(out.view(batch_size, S, S, T), x, forcing, v, t_interval)
+ loss_ic, loss_f = PINO_loss3d(
+ out.view(batch_size, S, S, T), x, forcing, v, t_interval
+ )
else:
loss_ic, loss_f = zero, zero
@@ -70,79 +77,84 @@ def train(model,
total_loss.backward()
optimizer.step()
- loss_dict['train_ic'] += loss_ic
- loss_dict['test_l2'] += loss_l2
- loss_dict['train_loss'] += total_loss
- loss_dict['train_f'] += loss_f
+ loss_dict["train_ic"] += loss_ic
+ loss_dict["test_l2"] += loss_l2
+ loss_dict["train_loss"] += total_loss
+ loss_dict["train_f"] += loss_f
if rank == 0 and profile:
paddle.cuda.synchronize()
t2 = default_timer()
- log_dict['Time cost'] = t2 - t1
+ log_dict["Time cost"] = t2 - t1
scheduler.step()
loss_reduced = reduce_loss_dict(loss_dict)
- train_ic = loss_reduced['train_ic'].item() / len(train_loader)
- train_f = loss_reduced['train_f'].item() / len(train_loader)
- train_loss = loss_reduced['train_loss'].item() / len(train_loader)
- test_l2 = loss_reduced['test_l2'].item() / len(train_loader)
+ train_ic = loss_reduced["train_ic"].item() / len(train_loader)
+ train_f = loss_reduced["train_f"].item() / len(train_loader)
+ train_loss = loss_reduced["train_loss"].item() / len(train_loader)
+ test_l2 = loss_reduced["test_l2"].item() / len(train_loader)
log_dict = {
- 'Train f error': train_f,
- 'Train L2 error': train_ic,
- 'Train loss': train_loss,
- 'Test L2 error': test_l2
- }
+ "Train f error": train_f,
+ "Train L2 error": train_ic,
+ "Train loss": train_loss,
+ "Test L2 error": test_l2,
+ }
if rank == 0:
if use_tqdm:
pbar.set_description(
(
- f'Train f error: {train_f:.5f}; Train ic l2 error: {train_ic:.5f}. '
- f'Train loss: {train_loss:.5f}; Test l2 error: {test_l2:.5f}'
+ f"Train f error: {train_f:.5f}; Train ic l2 error: {train_ic:.5f}. "
+ f"Train loss: {train_loss:.5f}; Test l2 error: {test_l2:.5f}"
)
)
if rank == 0:
- save_checkpoint(config['train']['save_dir'],
- config['train']['save_name'],
- model, optimizer)
-
-def mixed_train(model, # model of neural operator
- train_loader, # dataloader for training with data
- S1, T1, # spacial and time dimension for training with data
- a_loader, # generator for ICs
- S2, T2, # spacial and time dimension for training with equation only
- optimizer, # optimizer
- scheduler, # learning rate scheduler
- config, # configuration dict
- log=False, # turn on the wandb
- project='PINO-default', # project name
- group='FDM', # group name
- tags=['Nan'], # tags
- use_tqdm=True): # turn on tqdm
+ save_checkpoint(
+ config["train"]["save_dir"], config["train"]["save_name"], model, optimizer
+ )
+
+
+def mixed_train(
+ model, # model of neural operator
+ train_loader, # dataloader for training with data
+ S1,
+ T1, # spacial and time dimension for training with data
+ a_loader, # generator for ICs
+ S2,
+ T2, # spacial and time dimension for training with equation only
+ optimizer, # optimizer
+ scheduler, # learning rate scheduler
+ config, # configuration dict
+ log=False, # turn on the wandb
+ project="PINO-default", # project name
+ group="FDM", # group name
+ tags=["Nan"], # tags
+ use_tqdm=True,
+): # turn on tqdm
# data parameters
- v = 1 / config['data']['Re']
- t_interval = config['data']['time_interval']
+ v = 1 / config["data"]["Re"]
+ t_interval = config["data"]["time_interval"]
forcing_1 = get_forcing(S1)
forcing_2 = get_forcing(S2)
# training settings
- batch_size = config['train']['batchsize']
- ic_weight = config['train']['ic_loss']
- f_weight = config['train']['f_loss']
- xy_weight = config['train']['xy_loss']
- num_data_iter = config['train']['data_iter']
- num_eqn_iter = config['train']['eqn_iter']
+ batch_size = config["train"]["batchsize"]
+ ic_weight = config["train"]["ic_loss"]
+ f_weight = config["train"]["f_loss"]
+ xy_weight = config["train"]["xy_loss"]
+ num_data_iter = config["train"]["data_iter"]
+ num_eqn_iter = config["train"]["eqn_iter"]
model.train()
myloss = LpLoss(size_average=True)
- pbar = range(config['train']['epochs'])
+ pbar = range(config["train"]["epochs"])
if use_tqdm:
pbar = tqdm(pbar, dynamic_ncols=True, smoothing=0.05)
- zero = paddle.zeros(1, dtype='float32')
+ zero = paddle.zeros(1, dtype="float32")
train_loader = sample_data(train_loader)
for ep in pbar:
model.train()
- t1 = default_timer()
+ default_timer()
train_loss = 0.0
train_ic = 0.0
train_f = 0.0
@@ -158,13 +170,15 @@ def mixed_train(model, # model of neural operator
out = out[..., :-5]
x = x[:, :, :, 0, -1]
- loss_l2 = myloss(out.reshape([batch_size, S1, S1, T1]),
- y.reshape([batch_size, S1, S1, T1]))
+ loss_l2 = myloss(
+ out.reshape([batch_size, S1, S1, T1]),
+ y.reshape([batch_size, S1, S1, T1]),
+ )
if ic_weight != 0 or f_weight != 0:
- loss_ic, loss_f = PINO_loss3d(out.reshape([batch_size, S1, S1, T1]),
- x, forcing_1,
- v, t_interval)
+ loss_ic, loss_f = PINO_loss3d(
+ out.reshape([batch_size, S1, S1, T1]), x, forcing_1, v, t_interval
+ )
else:
loss_ic, loss_f = zero, zero
@@ -191,9 +205,9 @@ def mixed_train(model, # model of neural operator
out = model(x_in).reshape([batch_size, S2, S2, T2 + 5])
out = out[..., :-5]
new_a = new_a[:, :, :, 0, -1]
- loss_ic, loss_f = PINO_loss3d(out.reshape([batch_size, S2, S2, T2]),
- new_a, forcing_2,
- v, t_interval)
+ loss_ic, loss_f = PINO_loss3d(
+ out.reshape([batch_size, S2, S2, T2]), new_a, forcing_2, v, t_interval
+ )
eqn_loss = loss_f * f_weight + loss_ic * ic_weight
eqn_loss.backward()
optimizer.step()
@@ -201,56 +215,62 @@ def mixed_train(model, # model of neural operator
err_eqn += eqn_loss.item()
scheduler.step()
- t2 = default_timer()
+ default_timer()
if num_eqn_iter != 0:
err_eqn /= num_eqn_iter
if use_tqdm:
pbar.set_description(
(
- f'Data f error: {train_f:.5f}; Data ic l2 error: {train_ic:.5f}. '
- f'Data train loss: {train_loss:.5f}; Data l2 error: {test_l2:.5f}'
- f'Eqn loss: {err_eqn:.5f}'
+ f"Data f error: {train_f:.5f}; Data ic l2 error: {train_ic:.5f}. "
+ f"Data train loss: {train_loss:.5f}; Data l2 error: {test_l2:.5f}"
+ f"Eqn loss: {err_eqn:.5f}"
)
)
- save_checkpoint(config['train']['save_dir'],
- config['train']['save_name'],
- model, optimizer)
-
-def progressive_train(model,
- loader, train_loader,
- optimizer, scheduler,
- milestones, config,
- log=False,
- project='PINO-default',
- group='FDM',
- tags=['Nan'],
- use_tqdm=True):
+ save_checkpoint(
+ config["train"]["save_dir"], config["train"]["save_name"], model, optimizer
+ )
+
+
+def progressive_train(
+ model,
+ loader,
+ train_loader,
+ optimizer,
+ scheduler,
+ milestones,
+ config,
+ log=False,
+ project="PINO-default",
+ group="FDM",
+ tags=["Nan"],
+ use_tqdm=True,
+):
# data parameters
- v = 1 / config['data']['Re']
+ v = 1 / config["data"]["Re"]
T = loader.T
- t_interval = config['data']['time_interval']
+ t_interval = config["data"]["time_interval"]
# training settings
- batch_size = config['train']['batchsize']
- ic_weight = config['train']['ic_loss']
- f_weight = config['train']['f_loss']
- xy_weight = config['train']['xy_loss']
+ batch_size = config["train"]["batchsize"]
+ ic_weight = config["train"]["ic_loss"]
+ f_weight = config["train"]["f_loss"]
+ xy_weight = config["train"]["xy_loss"]
model.train()
myloss = LpLoss(size_average=True)
- zero = paddle.zeros(1, dtype='float32')
- for milestone, epochs in zip(milestones, config['train']['epochs']):
+ zero = paddle.zeros(1, dtype="float32")
+ for milestone, epochs in zip(milestones, config["train"]["epochs"]):
pbar = range(epochs)
if use_tqdm:
pbar = tqdm(pbar, dynamic_ncols=True, smoothing=0.05)
S = loader.S // milestone
- print(f'Resolution :{S}')
+ print(f"Resolution :{S}")
forcing = get_forcing(S)
for ep in pbar:
model.train()
- t1 = default_timer()
+ default_timer()
train_loss = 0.0
train_ic = 0.0
train_f = 0.0
@@ -265,15 +285,20 @@ def progressive_train(model,
out = out[..., :-5]
x = x[:, :, :, 0, -1]
- loss_l2 = myloss(out.view(batch_size, S, S, T), y.view(batch_size, S, S, T))
+ loss_l2 = myloss(
+ out.view(batch_size, S, S, T), y.view(batch_size, S, S, T)
+ )
if ic_weight != 0 or f_weight != 0:
- loss_ic, loss_f = PINO_loss3d(out.view(batch_size, S, S, T),
- x, forcing, v, t_interval)
+ loss_ic, loss_f = PINO_loss3d(
+ out.view(batch_size, S, S, T), x, forcing, v, t_interval
+ )
else:
loss_ic, loss_f = zero, zero
- total_loss = loss_l2 * xy_weight + loss_f * f_weight + loss_ic * ic_weight
+ total_loss = (
+ loss_l2 * xy_weight + loss_f * f_weight + loss_ic * ic_weight
+ )
total_loss.backward()
@@ -288,16 +313,15 @@ def progressive_train(model,
train_f /= len(train_loader)
train_loss /= len(train_loader)
test_l2 /= len(train_loader)
- t2 = default_timer()
+ default_timer()
if use_tqdm:
pbar.set_description(
(
- f'Train f error: {train_f:.5f}; Train ic l2 error: {train_ic:.5f}. '
- f'Train loss: {train_loss:.5f}; Test l2 error: {test_l2:.5f}'
+ f"Train f error: {train_f:.5f}; Train ic l2 error: {train_ic:.5f}. "
+ f"Train loss: {train_loss:.5f}; Test l2 error: {test_l2:.5f}"
)
)
- save_checkpoint(config['train']['save_dir'],
- config['train']['save_name'],
- model, optimizer)
-
+ save_checkpoint(
+ config["train"]["save_dir"], config["train"]["save_name"], model, optimizer
+ )
diff --git a/jointContribution/PINO/PINO_paddle/train_utils/utils.py b/jointContribution/PINO/PINO_paddle/train_utils/utils.py
index 7b116aeb69..9dd352a30f 100644
--- a/jointContribution/PINO/PINO_paddle/train_utils/utils.py
+++ b/jointContribution/PINO/PINO_paddle/train_utils/utils.py
@@ -1,16 +1,18 @@
import os
+
import numpy as np
import paddle
+
def vor2vel(w, L=2 * np.pi):
- '''
+ """
Convert vorticity into velocity
Args:
w: vorticity with shape (batchsize, num_x, num_y, num_t)
Returns:
ux, uy with the same shape
- '''
+ """
batchsize = w.size(0)
nx = w.size(1)
ny = w.size(2)
@@ -22,107 +24,157 @@ def vor2vel(w, L=2 * np.pi):
# Wavenumbers in y-direction
k_max = nx // 2
N = nx
- k_x = paddle.cat((paddle.arange(start=0, end=k_max, step=1, device=device),
- paddle.arange(start=-k_max, end=0, step=1, device=device)), 0) \
- .reshape(N, 1).repeat(1, N).reshape(1, N, N, 1)
- k_y = paddle.cat((paddle.arange(start=0, end=k_max, step=1, device=device),
- paddle.arange(start=-k_max, end=0, step=1, device=device)), 0) \
- .reshape(1, N).repeat(N, 1).reshape(1, N, N, 1)
+ k_x = (
+ paddle.cat(
+ (
+ paddle.arange(start=0, end=k_max, step=1, device=device),
+ paddle.arange(start=-k_max, end=0, step=1, device=device),
+ ),
+ 0,
+ )
+ .reshape(N, 1)
+ .repeat(1, N)
+ .reshape(1, N, N, 1)
+ )
+ k_y = (
+ paddle.cat(
+ (
+ paddle.arange(start=0, end=k_max, step=1, device=device),
+ paddle.arange(start=-k_max, end=0, step=1, device=device),
+ ),
+ 0,
+ )
+ .reshape(1, N)
+ .repeat(N, 1)
+ .reshape(1, N, N, 1)
+ )
# Negative Laplacian in Fourier space
- lap = (k_x ** 2 + k_y ** 2)
+ lap = k_x**2 + k_y**2
lap[0, 0, 0, 0] = 1.0
f_h = w_h / lap
ux_h = 2 * np.pi / L * 1j * k_y * f_h
uy_h = -2 * np.pi / L * 1j * k_x * f_h
- ux = paddle.fft.irfft2(ux_h[:, :, :k_max + 1], dim=[1, 2])
- uy = paddle.fft.irfft2(uy_h[:, :, :k_max + 1], dim=[1, 2])
+ ux = paddle.fft.irfft2(ux_h[:, :, : k_max + 1], dim=[1, 2])
+ uy = paddle.fft.irfft2(uy_h[:, :, : k_max + 1], dim=[1, 2])
return ux, uy
+
def get_sample(N, T, s, p, q):
# sample p nodes from Initial Condition, p nodes from Boundary Condition, q nodes from Interior
# sample IC
index_ic = paddle.randint(s, size=(N, p))
sample_ic_t = paddle.zeros(N, p)
- sample_ic_x = index_ic/s
+ sample_ic_x = index_ic / s
# sample BC
- sample_bc = paddle.rand(size=(N, p//2))
- sample_bc_t = paddle.cat([sample_bc, sample_bc],dim=1)
- sample_bc_x = paddle.cat([paddle.zeros(N, p//2), paddle.ones(N, p//2)],dim=1)
+ sample_bc = paddle.rand(size=(N, p // 2))
+ sample_bc_t = paddle.cat([sample_bc, sample_bc], dim=1)
+ sample_bc_x = paddle.cat([paddle.zeros(N, p // 2), paddle.ones(N, p // 2)], dim=1)
- sample_i_t = -paddle.cos(paddle.rand(size=(N, q))*np.pi/2) + 1
- sample_i_x = paddle.rand(size=(N,q))
+ sample_i_t = -paddle.cos(paddle.rand(size=(N, q)) * np.pi / 2) + 1
+ sample_i_x = paddle.rand(size=(N, q))
sample_t = paddle.cat([sample_ic_t, sample_bc_t, sample_i_t], dim=1).cuda()
sample_t.requires_grad = True
sample_x = paddle.cat([sample_ic_x, sample_bc_x, sample_i_x], dim=1).cuda()
sample_x.requires_grad = True
- sample = paddle.stack([sample_t, sample_x], dim=-1).reshape(N, (p+p+q), 2)
+ sample = paddle.stack([sample_t, sample_x], dim=-1).reshape(N, (p + p + q), 2)
return sample, sample_t, sample_x, index_ic.long()
+
def get_grid(N, T, s):
- gridt = paddle.tensor(np.linspace(0, 1, T), dtype=paddle.float).reshape(1, T, 1).repeat(N, 1, s).cuda()
+ gridt = (
+ paddle.tensor(np.linspace(0, 1, T), dtype=paddle.float)
+ .reshape(1, T, 1)
+ .repeat(N, 1, s)
+ .cuda()
+ )
gridt.requires_grad = True
- gridx = paddle.tensor(np.linspace(0, 1, s+1)[:-1], dtype=paddle.float).reshape(1, 1, s).repeat(N, T, 1).cuda()
+ gridx = (
+ paddle.tensor(np.linspace(0, 1, s + 1)[:-1], dtype=paddle.float)
+ .reshape(1, 1, s)
+ .repeat(N, T, 1)
+ .cuda()
+ )
gridx.requires_grad = True
- grid = paddle.stack([gridt, gridx], dim=-1).reshape(N, T*s, 2)
+ grid = paddle.stack([gridt, gridx], dim=-1).reshape(N, T * s, 2)
return grid, gridt, gridx
+
def get_2dgrid(S):
- '''
+ """
get array of points on 2d grid in (0,1)^2
Args:
S: resolution
Returns:
points: flattened grid, ndarray (N, 2)
- '''
+ """
xarr = np.linspace(0, 1, S)
yarr = np.linspace(0, 1, S)
- xx, yy = np.meshgrid(xarr, yarr, indexing='ij')
+ xx, yy = np.meshgrid(xarr, yarr, indexing="ij")
points = np.stack([xx.ravel(), yy.ravel()], axis=0).T
return points
-def paddle2dgrid(num_x, num_y, bot=(0,0), top=(1,1)):
+
+def paddle2dgrid(num_x, num_y, bot=(0, 0), top=(1, 1)):
x_bot, y_bot = bot
x_top, y_top = top
x_arr = paddle.linspace(x_bot, x_top, num=num_x)
y_arr = paddle.linspace(y_bot, y_top, num=num_y)
- xx, yy = paddle.meshgrid(x_arr, y_arr, indexing='ij')
+ xx, yy = paddle.meshgrid(x_arr, y_arr, indexing="ij")
mesh = paddle.stack([xx, yy], dim=2)
return mesh
-def get_grid3d(S, T, time_scale=1.0, device='cpu'):
- gridx = paddle.tensor(np.linspace(0, 1, S + 1)[:-1], dtype=paddle.float, device=device)
+
+def get_grid3d(S, T, time_scale=1.0, device="cpu"):
+ gridx = paddle.tensor(
+ np.linspace(0, 1, S + 1)[:-1], dtype=paddle.float, device=device
+ )
gridx = gridx.reshape(1, S, 1, 1, 1).repeat([1, 1, S, T, 1])
- gridy = paddle.tensor(np.linspace(0, 1, S + 1)[:-1], dtype=paddle.float, device=device)
+ gridy = paddle.tensor(
+ np.linspace(0, 1, S + 1)[:-1], dtype=paddle.float, device=device
+ )
gridy = gridy.reshape(1, 1, S, 1, 1).repeat([1, S, 1, T, 1])
- gridt = paddle.tensor(np.linspace(0, 1 * time_scale, T), dtype=paddle.float, device=device)
+ gridt = paddle.tensor(
+ np.linspace(0, 1 * time_scale, T), dtype=paddle.float, device=device
+ )
gridt = gridt.reshape(1, 1, 1, T, 1).repeat([1, S, S, 1, 1])
return gridx, gridy, gridt
+
def convert_ic(u0, N, S, T, time_scale=1.0):
u0 = u0.reshape(N, S, S, 1, 1).repeat([1, 1, 1, T, 1])
gridx, gridy, gridt = get_grid3d(S, T, time_scale=time_scale, device=u0.device)
- a_data = paddle.cat((gridx.repeat([N, 1, 1, 1, 1]), gridy.repeat([N, 1, 1, 1, 1]),
- gridt.repeat([N, 1, 1, 1, 1]), u0), dim=-1)
+ a_data = paddle.cat(
+ (
+ gridx.repeat([N, 1, 1, 1, 1]),
+ gridy.repeat([N, 1, 1, 1, 1]),
+ gridt.repeat([N, 1, 1, 1, 1]),
+ u0,
+ ),
+ dim=-1,
+ )
return a_data
+
def requires_grad(model, flag=True):
for p in model.parameters():
p.requires_grad = flag
+
def set_grad(tensors, flag=True):
for p in tensors:
p.requires_grad = flag
+
def zero_grad(params):
- '''
+ """
set grad field to 0
- '''
+ """
if isinstance(params, paddle.Tensor):
if params.grad is not None:
params.grad.zero_()
@@ -131,14 +183,16 @@ def zero_grad(params):
if p.grad is not None:
p.grad.zero_()
+
def count_params(net):
count = 0
for p in net.parameters():
count += p.numel()
return count
+
def save_checkpoint(path, name, model, optimizer=None):
- ckpt_dir = 'checkpoints/%s/' % path
+ ckpt_dir = "checkpoints/%s/" % path
if not os.path.exists(ckpt_dir):
os.makedirs(ckpt_dir)
try:
@@ -151,11 +205,9 @@ def save_checkpoint(path, name, model, optimizer=None):
else:
optim_dict = 0.0
- paddle.save({
- 'model': model_state_dict,
- 'optim': optim_dict
- }, ckpt_dir + name)
- print('Checkpoint is saved at %s' % ckpt_dir + name)
+ paddle.save({"model": model_state_dict, "optim": optim_dict}, ckpt_dir + name)
+ print("Checkpoint is saved at %s" % ckpt_dir + name)
+
def save_ckpt(path, model, optimizer=None, scheduler=None):
model_state = model.state_dict()
@@ -163,20 +215,19 @@ def save_ckpt(path, model, optimizer=None, scheduler=None):
optim_state = optimizer.state_dict()
else:
optim_state = None
-
+
if scheduler:
scheduler_state = scheduler.state_dict()
else:
scheduler_state = None
- paddle.save({
- 'model': model_state,
- 'optim': optim_state,
- 'scheduler': scheduler_state
- }, path)
- print(f'Checkpoint is saved to {path}')
+ paddle.save(
+ {"model": model_state, "optim": optim_state, "scheduler": scheduler_state}, path
+ )
+ print(f"Checkpoint is saved to {path}")
+
def dict2str(log_dict):
- res = ''
+ res = ""
for key, value in log_dict.items():
- res += f'{key}: {value}|'
- return res
\ No newline at end of file
+ res += f"{key}: {value}|"
+ return res
diff --git a/jointContribution/PINO/rfcs/PINO.md b/jointContribution/PINO/rfcs/PINO.md
index 345896a5ef..2b13409440 100644
--- a/jointContribution/PINO/rfcs/PINO.md
+++ b/jointContribution/PINO/rfcs/PINO.md
@@ -56,7 +56,7 @@ class FNO2d(nn.Layer):
width=64, fc_dim=128,
layers=None,
in_dim=3, out_dim=1,
- act='gelu',
+ act='gelu',
pad_ratio=[0., 0.]):
super(FNO2d, self).__init__()
@@ -127,4 +127,4 @@ def FDM_Burgers(u, v, D=1):
- 202308 : 调研
- 202309 :基于Paddle API的复现
-- 202310 :整理项目产出,撰写案例文档
\ No newline at end of file
+- 202310 :整理项目产出,撰写案例文档
diff --git a/jointContribution/XPINNs/requirements.txt b/jointContribution/XPINNs/requirements.txt
index 26a2b438ff..74fa65e6b5 100644
--- a/jointContribution/XPINNs/requirements.txt
+++ b/jointContribution/XPINNs/requirements.txt
@@ -1,3 +1,3 @@
matplotlib
-scipy
numpy
+scipy
diff --git a/jointContribution/graphGalerkin/LinearElasticity.py b/jointContribution/graphGalerkin/LinearElasticity.py
index 333cb5f454..77dd620654 100644
--- a/jointContribution/graphGalerkin/LinearElasticity.py
+++ b/jointContribution/graphGalerkin/LinearElasticity.py
@@ -195,7 +195,7 @@ def hard_way(self):
ax1.set_title("FEM solution")
fig.tight_layout(pad=3.0)
- idx_xcg = [
+ [
i
for i in range(xcg.shape[1])
if 2 * i not in dbc_idx and 2 * i + 1 not in dbc_idx
@@ -264,7 +264,6 @@ def main_square(self):
lims = np.asarray([[0, 1], [0, 1]])
nel = [2, 2]
porder = 2
- nf = 4
msh = mesh_hcube(etype, lims, nel, porder).getmsh()
xcg = msh.xcg
e2vcg = msh.e2vcg
@@ -338,7 +337,7 @@ def main_square(self):
femsp_gcnn.spmat,
dbc,
)
- fcn_fem = lambda u_: create_fem_resjac(
+ lambda u_: create_fem_resjac(
"cg",
u_,
msh.transfdatacontiguous,
diff --git a/jointContribution/graphGalerkin/NsChebnet.py b/jointContribution/graphGalerkin/NsChebnet.py
index 4b2d4ecf3c..aa38ec4a29 100644
--- a/jointContribution/graphGalerkin/NsChebnet.py
+++ b/jointContribution/graphGalerkin/NsChebnet.py
@@ -1,4 +1,3 @@
-import pdb
import sys
import matplotlib.pyplot as plt
diff --git a/jointContribution/graphGalerkin/rfcs/graphGalerkin.md b/jointContribution/graphGalerkin/rfcs/graphGalerkin.md
index c969876d56..a5025ce992 100644
--- a/jointContribution/graphGalerkin/rfcs/graphGalerkin.md
+++ b/jointContribution/graphGalerkin/rfcs/graphGalerkin.md
@@ -22,7 +22,7 @@ PaddlePaddle目前无相关模型实现。
## 3.1 解决的问题
本项目基于嵌入物理信息的图神经网络求解偏微分方程,方程主要以以下形式出现:
$$
-\nabla \cdot F(u, \nabla u; \boldsymbol\mu) = S(u, \nabla u; \boldsymbol\mu) \: in \: \Omega
+\nabla \cdot F(u, \nabla u; \boldsymbol\mu) = S(u, \nabla u; \boldsymbol\mu) \: in \: \Omega
$$
边界条件为:
$$
@@ -78,4 +78,4 @@ $$
- 202307 : 调研
- 202308 :基于Paddle API的复现
-- 202309 :整理项目产出,撰写案例文档
\ No newline at end of file
+- 202309 :整理项目产出,撰写案例文档
diff --git a/jointContribution/graphGalerkin/source/FEM_ForwardModel.py b/jointContribution/graphGalerkin/source/FEM_ForwardModel.py
index c3cbbb7ce7..47cff78b4c 100644
--- a/jointContribution/graphGalerkin/source/FEM_ForwardModel.py
+++ b/jointContribution/graphGalerkin/source/FEM_ForwardModel.py
@@ -1,18 +1,11 @@
import numpy as np
-import pdb
-import matplotlib.pyplot as plt
-from pyCaMOtk.create_mesh_hsphere import mesh_hsphere
-from pyCaMOtk.setup_linelptc_sclr_base_handcode import setup_linelptc_sclr_base_handcode
-from pyCaMOtk.create_dbc_strct import create_dbc_strct
-from pyCaMOtk.create_femsp_cg import create_femsp_cg
-from pyCaMOtk.solve_fem import solve_fem
-from pyCaMOtk.visualize_fem import visualize_fem
-def analyticalPossion(xcg,Tc,Tb=0):
- Ue=Tc*(1-xcg[0,:]**2-xcg[1,:]**2)/4+Tb
+def analyticalPossion(xcg, Tc, Tb=0):
+ Ue = Tc * (1 - xcg[0, :] ** 2 - xcg[1, :] ** 2) / 4 + Tb
return Ue.flatten()
-def analyticalConeInterpolation(xcg,Tc,Tb=0):
- Ue=Tc*(1-np.sqrt(xcg[0,:]**2+xcg[1,:]**2))/4+Tb
- return Ue.flatten()
\ No newline at end of file
+
+def analyticalConeInterpolation(xcg, Tc, Tb=0):
+ Ue = Tc * (1 - np.sqrt(xcg[0, :] ** 2 + xcg[1, :] ** 2)) / 4 + Tb
+ return Ue.flatten()
diff --git a/jointContribution/graphGalerkin/source/GCNNModel.py b/jointContribution/graphGalerkin/source/GCNNModel.py
index f7b8c54fd5..726bb1d01b 100644
--- a/jointContribution/graphGalerkin/source/GCNNModel.py
+++ b/jointContribution/graphGalerkin/source/GCNNModel.py
@@ -1,13 +1,14 @@
# from pgl.nn import GCNConv
+import sys
+
import numpy as np
import paddle
import paddle.nn.initializer as Initializer
-import sys
sys.path.insert(0, "utils")
from ChebConv import ChebConv
+from paddle.nn import Layer
from paddle.nn.functional import relu
-from paddle.nn import Layer, Linear
place = paddle.CUDAPlace(0)
diff --git a/jointContribution/graphGalerkin/source/TensorFEMCore.py b/jointContribution/graphGalerkin/source/TensorFEMCore.py
index 9815510621..b23cb7b3cc 100644
--- a/jointContribution/graphGalerkin/source/TensorFEMCore.py
+++ b/jointContribution/graphGalerkin/source/TensorFEMCore.py
@@ -1,5 +1,4 @@
import os
-import pdb
import time
import matplotlib.pyplot as plt
@@ -91,7 +90,7 @@ def create_fem_resjac(
else:
raise ValueError("FE space only support cg!")
R = assemble_nobc_vec(Re, ldof2gdof_eqn)
- if enforce_idx == None:
+ if enforce_idx is None:
Rf = R[free_idx]
else:
Rf = R[enforce_idx]
@@ -128,7 +127,7 @@ def intg_elem_claw_vol(Ue, transf_data, elem, elem_data, e, parsfuncI=None, mode
[nvar_per_elem, nvar * (ndim + 1)], order="F"
)
x = transf_data.xq[:, k, e]
- if parsfuncI == None:
+ if parsfuncI is None:
pars = elem_data.vol_pars[:, k, e]
else:
pars = parsfuncI(x)
@@ -178,7 +177,7 @@ def intg_elem_claw_extface(Ue, transf_data, elem, elem_data, e, parsfuncB=None):
)
Teqn = Double(Teqn)
Tvar = Double(Tvar)
- if parsfuncB == None:
+ if parsfuncB is None:
pars = elem_data.bnd_pars[:, k, f, e]
else:
pars = parsfuncB(x)
@@ -207,7 +206,7 @@ def assemble_nobc_mat(Me, cooidx, lmat2gmat):
def assemble_nobc_vec(Fe, ldof2gdof_eqn):
"""Assembly global residual of conservation law (!!very useful!!)"""
ndof = np.max(ldof2gdof_eqn[:]) + 1
- nelem = Fe.shape[1]
+ Fe.shape[1]
F = np.zeros(shape=[ndof, 1])
F = Double(F)
F.stop_gradient = False
@@ -326,7 +325,6 @@ def trainmodel(
):
model.train()
er_0 = 0
- loss_0 = 0
erlist = []
ReList = []
optimizer.clear_grad()
diff --git a/jointContribution/graphGalerkin/source/setup_prob_eqn_handcode.py b/jointContribution/graphGalerkin/source/setup_prob_eqn_handcode.py
index c5c3e83a56..5566956e26 100644
--- a/jointContribution/graphGalerkin/source/setup_prob_eqn_handcode.py
+++ b/jointContribution/graphGalerkin/source/setup_prob_eqn_handcode.py
@@ -1,228 +1,261 @@
-import paddle
import numpy as np
-import pdb
-
-from TensorFEMCore import Double, ReshapeFix
+import paddle
+from TensorFEMCore import Double
+from TensorFEMCore import ReshapeFix
"""
####Possion Equation
"""
+
+
class setup_linelptc_sclr_base_handcode(object):
- """docstring for setup_linelptc_sclr_base_handcode"""
- def __init__(self,ndim,K,f,Qb,bnd2nbc):
- self.ndim=ndim
- self.K=K
- self.f=f
- self.Qb=Qb
- self.bnd2nbc=bnd2nbc
-
- self.I=np.eye(self.ndim)
- if self.K==None:
- self.K=lambda x,el: self.I.reshape(self.ndim**2,1,order='F') #Fortan like
- if self.f==None:
- self.f=lambda x,el: 0
- if self.Qb==None:
- self.Qb=lambda x,n,bnd,el,fc: 0
-
- self.eqn=LinearEllipticScalarBaseHandcode()
- self.vol_pars_fcn=lambda x,el:np.vstack((self.K(x, el),self.f(x, el),np.nan))
- self.bnd_pars_fcn=lambda x,n,bnd,el,fc:np.vstack((self.K(x,el),
- self.f(x,el),
- self.Qb(x,n,bnd,el,fc)))
-
-
+ """docstring for setup_linelptc_sclr_base_handcode"""
+
+ def __init__(self, ndim, K, f, Qb, bnd2nbc):
+ self.ndim = ndim
+ self.K = K
+ self.f = f
+ self.Qb = Qb
+ self.bnd2nbc = bnd2nbc
+
+ self.I = np.eye(self.ndim)
+ if self.K is None:
+ self.K = lambda x, el: self.I.reshape(
+ self.ndim**2, 1, order="F"
+ ) # Fortan like
+ if self.f is None:
+ self.f = lambda x, el: 0
+ if self.Qb is None:
+ self.Qb = lambda x, n, bnd, el, fc: 0
+
+ self.eqn = LinearEllipticScalarBaseHandcode()
+ self.vol_pars_fcn = lambda x, el: np.vstack(
+ (self.K(x, el), self.f(x, el), np.nan)
+ )
+ self.bnd_pars_fcn = lambda x, n, bnd, el, fc: np.vstack(
+ (self.K(x, el), self.f(x, el), self.Qb(x, n, bnd, el, fc))
+ )
+
class LinearEllipticScalarBaseHandcode(object):
- """docstring for LinearEllipticScalarBaseHandcode"""
- def __init__(self):
- self.neqn=1
- self.nvar=1
- self.ncomp=1
-
- def srcflux(self,UQ,pars,x,model=None):
- """
- eval_linelptc_base_handcode_srcflux
- """
- # Extract information from input
- q=UQ[0,1:]
- q=ReshapeFix(q,[len(q),1])
- self.ndim=len(q)
- try:
- k=np.reshape(pars[0:self.ndim**2],
- (self.ndim,self.ndim),order='F')
- except:
- k=paddle.reshape(pars[0:self.ndim**2],
- (self.ndim,self.ndim))
- f=pars[self.ndim**2]
- try:
- temp_flag=(f.requires_grad)
- f=f.reshape([1,1])
- except:
- f=paddle.to_tensor(f, dtype='float32').reshape([1,1])
-
- k_ml=paddle.to_tensor(k, dtype='float32')
- # Define flux and source
- SF=paddle.concat((f,-1*paddle.mm(k_ml,q)),axis=0)
-
- # Define partial derivative
- dSFdU=np.zeros([self.neqn, self.ndim+1, self.ncomp,self.ndim+1])
- try:
- dSFdU[:,1:,:,1:]=np.reshape(-1*k,[self.neqn, self.ndim,self.ncomp,self.ndim])
- except:
- k=k.detach().cpu().numpy()
- dSFdU[:,1:,:,1:]=np.reshape(-1*k,[self.neqn, self.ndim,self.ncomp,self.ndim])
- dSFdU=paddle.to_tensor(dSFdU, dtype='float32')
- return SF, dSFdU
-
- def bndstvcflux(self,nbcnbr,UQ,pars,x,n):
- nvar=UQ.shape[0]
- ndim=UQ.shape[1]-1
-
- Ub=UQ[:,0]
- dUb=np.zeros([nvar,nvar,self.ndim+1])
- dUb[:,:,0]=np.eye(nvar)
-
- Fn=pars[ndim**2+1]
- dFn=np.zeros([nvar,nvar,self.ndim+1])
- dUb=Double(dUb)
- Fn=Double(Fn)
- dFn=Double(dFn)
- return Ub,dUb,Fn,dFn
+ """docstring for LinearEllipticScalarBaseHandcode"""
+
+ def __init__(self):
+ self.neqn = 1
+ self.nvar = 1
+ self.ncomp = 1
+
+ def srcflux(self, UQ, pars, x, model=None):
+ """
+ eval_linelptc_base_handcode_srcflux
+ """
+ # Extract information from input
+ q = UQ[0, 1:]
+ q = ReshapeFix(q, [len(q), 1])
+ self.ndim = len(q)
+ try:
+ k = np.reshape(pars[0 : self.ndim**2], (self.ndim, self.ndim), order="F")
+ except:
+ k = paddle.reshape(pars[0 : self.ndim**2], (self.ndim, self.ndim))
+ f = pars[self.ndim**2]
+ try:
+ f = f.reshape([1, 1])
+ except:
+ f = paddle.to_tensor(f, dtype="float32").reshape([1, 1])
+
+ k_ml = paddle.to_tensor(k, dtype="float32")
+ # Define flux and source
+ SF = paddle.concat((f, -1 * paddle.mm(k_ml, q)), axis=0)
+
+ # Define partial derivative
+ dSFdU = np.zeros([self.neqn, self.ndim + 1, self.ncomp, self.ndim + 1])
+ try:
+ dSFdU[:, 1:, :, 1:] = np.reshape(
+ -1 * k, [self.neqn, self.ndim, self.ncomp, self.ndim]
+ )
+ except:
+ k = k.detach().cpu().numpy()
+ dSFdU[:, 1:, :, 1:] = np.reshape(
+ -1 * k, [self.neqn, self.ndim, self.ncomp, self.ndim]
+ )
+ dSFdU = paddle.to_tensor(dSFdU, dtype="float32")
+ return SF, dSFdU
+
+ def bndstvcflux(self, nbcnbr, UQ, pars, x, n):
+ nvar = UQ.shape[0]
+ ndim = UQ.shape[1] - 1
+
+ Ub = UQ[:, 0]
+ dUb = np.zeros([nvar, nvar, self.ndim + 1])
+ dUb[:, :, 0] = np.eye(nvar)
+
+ Fn = pars[ndim**2 + 1]
+ dFn = np.zeros([nvar, nvar, self.ndim + 1])
+ dUb = Double(dUb)
+ Fn = Double(Fn)
+ dFn = Double(dFn)
+ return Ub, dUb, Fn, dFn
+
"""
####Linear Elasticity Equation
"""
+
class setup_linelast_base_handcode(object):
- """docstring for setup_linelast_base_handcode"""
- def __init__(self,ndim,lam,mu,f,tb,bnd2nbc):
- self.bnd2nbc=bnd2nbc
- self.eqn=LinearElasticityBaseHandcode(ndim)
- self.vol_pars_fcn=lambda x, el: np.vstack((lam(x,el),
- mu(x,el),
- f(x,el),
- np.zeros([ndim,1])+np.nan))
- self.bnd_pars_fcn=lambda x,n,bnd,el,fc:np.vstack((lam(x, el),
- mu(x, el),
- f(x, el),
- tb(x, n, bnd, el, fc)))
-
-
+ """docstring for setup_linelast_base_handcode"""
+
+ def __init__(self, ndim, lam, mu, f, tb, bnd2nbc):
+ self.bnd2nbc = bnd2nbc
+ self.eqn = LinearElasticityBaseHandcode(ndim)
+ self.vol_pars_fcn = lambda x, el: np.vstack(
+ (lam(x, el), mu(x, el), f(x, el), np.zeros([ndim, 1]) + np.nan)
+ )
+ self.bnd_pars_fcn = lambda x, n, bnd, el, fc: np.vstack(
+ (lam(x, el), mu(x, el), f(x, el), tb(x, n, bnd, el, fc))
+ )
+
class LinearElasticityBaseHandcode(object):
- """docstring for LinearElasticityBaseHandcode"""
- def __init__(self,ndim):
- self.neqn=ndim
- self.nvar=ndim
- self.bndstvcflux=\
- lambda nbcnbr, UQ, pars, x, n:\
- eval_linelast_base_handcode_bndstvc_intr_bndflux_pars(UQ, pars, x, n)
- self.srcflux=lambda UQ,pars,x:\
- eval_linelast_base_handcode_srcflux(UQ, pars, x)
+ """docstring for LinearElasticityBaseHandcode"""
+
+ def __init__(self, ndim):
+ self.neqn = ndim
+ self.nvar = ndim
+ self.bndstvcflux = lambda nbcnbr, UQ, pars, x, n: eval_linelast_base_handcode_bndstvc_intr_bndflux_pars(
+ UQ, pars, x, n
+ )
+ self.srcflux = lambda UQ, pars, x: eval_linelast_base_handcode_srcflux(
+ UQ, pars, x
+ )
+
def eval_linelast_base_handcode_srcflux(UQ, pars, x):
- q=UQ[:,1:]
- ndim=q.shape[0]
- # Define information regarding size of the system
- neqn=ndim
- ncomp=ndim
-
- # Extract parameters
- lam=pars[0]
- mu=pars[1]
- f=pars[2:2+ndim]
- F=-lam*paddle.trace(q)*(Double(np.eye(ndim)))-mu*(q+q.T)
- try:
- S=Double(f.reshape([ndim,1],order='F'))
- except:
- S=f.reshape([ndim,1])
- SF=paddle.concat((S,F),axis=1)
- dSFdU=Double(np.zeros([neqn,ndim+1,ncomp,ndim+1]))
- for i in range(ndim):
- for j in range(ndim):
- dSFdU[i,1+i,j,1+j]=dSFdU[i,1+i,j,1+j]-lam
- dSFdU[i,1+j,i,1+j]=dSFdU[i,1+j,i,1+j]-mu
- dSFdU[i,1+j,j,1+i]=dSFdU[i,1+j,j,1+i]-mu
- return SF, dSFdU
-
-def eval_linelast_base_handcode_bndstvc_intr_bndflux_pars(UQ,pars,x,n):
- nvar=UQ.shape[0]
- ndim=UQ.shape[1]-1
-
- Ub=UQ[:,0]
- dUb=np.zeros([nvar,nvar,ndim+1])
- dUb[:,:,0]=np.eye(nvar)
- Fn=-pars[-ndim:]
- dFn=np.zeros([nvar,nvar,ndim+1])
- dUb=Double(dUb)
- Fn=ReshapeFix(Double(Fn),[len(Fn),1],order='F')
- dFn=Double(dFn)
- return Ub,dUb,Fn,dFn
+ q = UQ[:, 1:]
+ ndim = q.shape[0]
+ # Define information regarding size of the system
+ neqn = ndim
+ ncomp = ndim
+
+ # Extract parameters
+ lam = pars[0]
+ mu = pars[1]
+ f = pars[2 : 2 + ndim]
+ F = -lam * paddle.trace(q) * (Double(np.eye(ndim))) - mu * (q + q.T)
+ try:
+ S = Double(f.reshape([ndim, 1], order="F"))
+ except:
+ S = f.reshape([ndim, 1])
+ SF = paddle.concat((S, F), axis=1)
+ dSFdU = Double(np.zeros([neqn, ndim + 1, ncomp, ndim + 1]))
+ for i in range(ndim):
+ for j in range(ndim):
+ dSFdU[i, 1 + i, j, 1 + j] = dSFdU[i, 1 + i, j, 1 + j] - lam
+ dSFdU[i, 1 + j, i, 1 + j] = dSFdU[i, 1 + j, i, 1 + j] - mu
+ dSFdU[i, 1 + j, j, 1 + i] = dSFdU[i, 1 + j, j, 1 + i] - mu
+ return SF, dSFdU
+
+
+def eval_linelast_base_handcode_bndstvc_intr_bndflux_pars(UQ, pars, x, n):
+ nvar = UQ.shape[0]
+ ndim = UQ.shape[1] - 1
+
+ Ub = UQ[:, 0]
+ dUb = np.zeros([nvar, nvar, ndim + 1])
+ dUb[:, :, 0] = np.eye(nvar)
+ Fn = -pars[-ndim:]
+ dFn = np.zeros([nvar, nvar, ndim + 1])
+ dUb = Double(dUb)
+ Fn = ReshapeFix(Double(Fn), [len(Fn), 1], order="F")
+ dFn = Double(dFn)
+ return Ub, dUb, Fn, dFn
+
"""
#### Inconpressible Navier Stokes Equation
"""
+
+
class setup_ins_base_handcode(object):
- """docstring for setup_ins_base_handcode"""
- def __init__(self,ndim,rho,nu,tb,bnd2nbc):
- self.eqn=IncompressibleNavierStokes(ndim)
- self.bnd2nbc=bnd2nbc
- self.vol_pars_fcn=lambda x,el:np.vstack([rho(x, el),
- nu(x, el),
- np.zeros([ndim+1,1])+np.nan])
- self.bnd_pars_fcn=lambda x,n,bnd,el,fc:np.vstack([rho(x,el),
- nu(x,el),
- tb(x,n,bnd,el,fc)])
+ """docstring for setup_ins_base_handcode"""
+
+ def __init__(self, ndim, rho, nu, tb, bnd2nbc):
+ self.eqn = IncompressibleNavierStokes(ndim)
+ self.bnd2nbc = bnd2nbc
+ self.vol_pars_fcn = lambda x, el: np.vstack(
+ [rho(x, el), nu(x, el), np.zeros([ndim + 1, 1]) + np.nan]
+ )
+ self.bnd_pars_fcn = lambda x, n, bnd, el, fc: np.vstack(
+ [rho(x, el), nu(x, el), tb(x, n, bnd, el, fc)]
+ )
+
class IncompressibleNavierStokes(object):
- """docstring for IncompressibleNavierStokes"""
- def __init__(self,ndim):
- self.ndim=ndim
- self.nvar=ndim+1
- self.srcflux=lambda UQ,pars,x:\
- eval_ins_base_handcode_srcflux(UQ,pars,x)
- self.bndstvcflux=lambda nbcnbr,UQ,pars,x,n:\
- eval_ins_base_handcode_bndstvc_intr_bndflux_pars(UQ,pars,x,n)
-
-def eval_ins_base_handcode_srcflux(UQ,pars,x):
- u=UQ[:,0]; q=UQ[:,1:]
- ndim=u.shape[0]-1
- neqn=ndim+1
- ncomp=ndim+1
- rho=pars[0]
- nu=pars[1]
- v=u[0:ndim]
-
- v=ReshapeFix(v,[len(v),1],'F')
-
- p=u[-1]
- dv=q[0:ndim,:]
- S=paddle.concat([-rho*paddle.mm(dv,v),-paddle.trace(dv).reshape([1,1])],axis=0)
-
- F=paddle.concat([-rho*nu*dv+p*paddle.eye(ndim, dtype='float32'),
- paddle.zeros([1,ndim], dtype='float32')],axis=0)
-
-
- SF=paddle.concat([S,F],axis=1)
-
- dSFdUQ=np.zeros([neqn,ndim+1,ncomp,ndim+1])
- dSFdUQ[:,0,:,0]=np.vstack([np.hstack([-rho*dv.detach().cpu().numpy(),np.zeros([ndim,1])]), np.zeros([1,ndim+1])])
- for i in range(ndim):
- dSFdUQ[i,0,i,1:]=-rho*v.detach().cpu().numpy().reshape(dSFdUQ[i,0,i,1:].shape,order='F')
- dSFdUQ[-1,0,0:-1,1:]=np.reshape(-np.eye(ndim),[1,ndim,ndim],order='F')
- dSFdUQ[0:-1,1:,-1,0]=np.eye(ndim)
- for i in range(ndim):
- for j in range(ndim):
- dSFdUQ[i,1+j,i,1+j]=dSFdUQ[i,1+j,i,1+j]-rho*nu
- dSFdUQ=Double(dSFdUQ)
- return SF,dSFdUQ
-
-def eval_ins_base_handcode_bndstvc_intr_bndflux_pars(UQ,pars,x,n):
- nvar=UQ.shape[0]
- ndim=UQ.shape[1]-1
- Ub=UQ[:,0]
- dUb=np.zeros([nvar,nvar,ndim+1])
- dUb[:,:,0]=np.eye(nvar)
- Fn=-pars[-ndim-1:].reshape([-1,1])
- dFn=np.zeros([nvar,nvar,ndim+1])
- return Ub,Double(dUb),Double(Fn),Double(dFn)
+ """docstring for IncompressibleNavierStokes"""
+
+ def __init__(self, ndim):
+ self.ndim = ndim
+ self.nvar = ndim + 1
+ self.srcflux = lambda UQ, pars, x: eval_ins_base_handcode_srcflux(UQ, pars, x)
+ self.bndstvcflux = lambda nbcnbr, UQ, pars, x, n: eval_ins_base_handcode_bndstvc_intr_bndflux_pars(
+ UQ, pars, x, n
+ )
+
+
+def eval_ins_base_handcode_srcflux(UQ, pars, x):
+ u = UQ[:, 0]
+ q = UQ[:, 1:]
+ ndim = u.shape[0] - 1
+ neqn = ndim + 1
+ ncomp = ndim + 1
+ rho = pars[0]
+ nu = pars[1]
+ v = u[0:ndim]
+
+ v = ReshapeFix(v, [len(v), 1], "F")
+
+ p = u[-1]
+ dv = q[0:ndim, :]
+ S = paddle.concat(
+ [-rho * paddle.mm(dv, v), -paddle.trace(dv).reshape([1, 1])], axis=0
+ )
+
+ F = paddle.concat(
+ [
+ -rho * nu * dv + p * paddle.eye(ndim, dtype="float32"),
+ paddle.zeros([1, ndim], dtype="float32"),
+ ],
+ axis=0,
+ )
+
+ SF = paddle.concat([S, F], axis=1)
+
+ dSFdUQ = np.zeros([neqn, ndim + 1, ncomp, ndim + 1])
+ dSFdUQ[:, 0, :, 0] = np.vstack(
+ [
+ np.hstack([-rho * dv.detach().cpu().numpy(), np.zeros([ndim, 1])]),
+ np.zeros([1, ndim + 1]),
+ ]
+ )
+ for i in range(ndim):
+ dSFdUQ[i, 0, i, 1:] = -rho * v.detach().cpu().numpy().reshape(
+ dSFdUQ[i, 0, i, 1:].shape, order="F"
+ )
+ dSFdUQ[-1, 0, 0:-1, 1:] = np.reshape(-np.eye(ndim), [1, ndim, ndim], order="F")
+ dSFdUQ[0:-1, 1:, -1, 0] = np.eye(ndim)
+ for i in range(ndim):
+ for j in range(ndim):
+ dSFdUQ[i, 1 + j, i, 1 + j] = dSFdUQ[i, 1 + j, i, 1 + j] - rho * nu
+ dSFdUQ = Double(dSFdUQ)
+ return SF, dSFdUQ
+
+
+def eval_ins_base_handcode_bndstvc_intr_bndflux_pars(UQ, pars, x, n):
+ nvar = UQ.shape[0]
+ ndim = UQ.shape[1] - 1
+ Ub = UQ[:, 0]
+ dUb = np.zeros([nvar, nvar, ndim + 1])
+ dUb[:, :, 0] = np.eye(nvar)
+ Fn = -pars[-ndim - 1 :].reshape([-1, 1])
+ dFn = np.zeros([nvar, nvar, ndim + 1])
+ return Ub, Double(dUb), Double(Fn), Double(dFn)
diff --git a/jointContribution/graphGalerkin/utils/init.py b/jointContribution/graphGalerkin/utils/init.py
index e7f7ba175d..3cbdebe676 100644
--- a/jointContribution/graphGalerkin/utils/init.py
+++ b/jointContribution/graphGalerkin/utils/init.py
@@ -1,74 +1,82 @@
-from typing import Any
-
import math
+from typing import Any
import paddle
from paddle import Tensor
from paddle.nn.initializer import Orthogonal
+
def uniform(size: int, value: Any):
if isinstance(value, Tensor):
bound = 1.0 / math.sqrt(size)
value.data.uniform_(-bound, bound)
else:
- for v in value.parameters() if hasattr(value, 'parameters') else []:
+ for v in value.parameters() if hasattr(value, "parameters") else []:
uniform(size, v)
- for v in value.buffers() if hasattr(value, 'buffers') else []:
+ for v in value.buffers() if hasattr(value, "buffers") else []:
uniform(size, v)
+
def kaiming_uniform(value: Any, fan: int, a: float):
if isinstance(value, Tensor):
bound = math.sqrt(6 / ((1 + a**2) * fan))
value.data.uniform_(-bound, bound)
else:
- for v in value.parameters() if hasattr(value, 'parameters') else []:
+ for v in value.parameters() if hasattr(value, "parameters") else []:
kaiming_uniform(v, fan, a)
- for v in value.buffers() if hasattr(value, 'buffers') else []:
+ for v in value.buffers() if hasattr(value, "buffers") else []:
kaiming_uniform(v, fan, a)
+
def glorot(value: Any):
if isinstance(value, Tensor):
stdv = math.sqrt(6.0 / (value.shape[-2] + value.shape[-1]))
value = paddle.uniform(value.shape, value.dtype, -stdv, stdv)
else:
- for v in value.parameters() if hasattr(value, 'parameters') else []:
+ for v in value.parameters() if hasattr(value, "parameters") else []:
glorot(v)
- for v in value.buffers() if hasattr(value, 'buffers') else []:
+ for v in value.buffers() if hasattr(value, "buffers") else []:
glorot(v)
+
def glorot_orthogonal(tensor, scale):
if tensor is not None:
tensor = paddle.create_parameter(tensor.shape, attr=Orthogonal())
- scale /= ((tensor.size(-2) + tensor.size(-1)) * tensor.var())
+ scale /= (tensor.size(-2) + tensor.size(-1)) * tensor.var()
tensor.data *= scale.sqrt()
+
def constant(value: Any, fill_value: float):
if isinstance(value, Tensor):
value = paddle.full(value.shape, fill_value, value.dtype)
else:
- for v in value.parameters() if hasattr(value, 'parameters') else []:
+ for v in value.parameters() if hasattr(value, "parameters") else []:
constant(v, fill_value)
- for v in value.buffers() if hasattr(value, 'buffers') else []:
+ for v in value.buffers() if hasattr(value, "buffers") else []:
constant(v, fill_value)
+
def zeros(value: Any):
- constant(value, 0.)
+ constant(value, 0.0)
+
def ones(tensor: Any):
- constant(tensor, 1.)
+ constant(tensor, 1.0)
+
def normal(value: Any, mean: float, std: float):
if isinstance(value, Tensor):
value.data.normal_(mean, std)
else:
- for v in value.parameters() if hasattr(value, 'parameters') else []:
+ for v in value.parameters() if hasattr(value, "parameters") else []:
normal(v, mean, std)
- for v in value.buffers() if hasattr(value, 'buffers') else []:
+ for v in value.buffers() if hasattr(value, "buffers") else []:
normal(v, mean, std)
+
def reset(value: Any):
- if hasattr(value, 'reset_parameters'):
+ if hasattr(value, "reset_parameters"):
value.reset_parameters()
else:
- for child in value.children() if hasattr(value, 'children') else []:
+ for child in value.children() if hasattr(value, "children") else []:
reset(child)
diff --git a/jointContribution/graphGalerkin/utils/inspector.py b/jointContribution/graphGalerkin/utils/inspector.py
index 249187a549..56dcc7c647 100644
--- a/jointContribution/graphGalerkin/utils/inspector.py
+++ b/jointContribution/graphGalerkin/utils/inspector.py
@@ -1,16 +1,20 @@
-import re
import inspect
+import re
from collections import OrderedDict
-from typing import Dict, List, Any, Optional, Callable, Set, Tuple
-import pyparsing as pp
+from typing import Any
+from typing import Callable
+from typing import Dict
+from typing import List
+from typing import Optional
+from typing import Set
+
class Inspector(object):
def __init__(self, base_class: Any):
self.base_class: Any = base_class
self.params: Dict[str, Dict[str, Any]] = {}
- def inspect(self, func: Callable,
- pop_first: bool = False) -> Dict[str, Any]:
+ def inspect(self, func: Callable, pop_first: bool = False) -> Dict[str, Any]:
params = inspect.signature(func).parameters
params = OrderedDict(params)
if pop_first:
@@ -24,7 +28,7 @@ def keys(self, func_names: Optional[List[str]] = None) -> Set[str]:
return set(keys)
def __implements__(self, cls, func_name: str) -> bool:
- if cls.__name__ == 'MessagePassing':
+ if cls.__name__ == "MessagePassing":
return False
if func_name in cls.__dict__.keys():
return True
@@ -39,30 +43,31 @@ def distribute(self, func_name, kwargs: Dict[str, Any]):
data = kwargs.get(key, inspect.Parameter.empty)
if data is inspect.Parameter.empty:
if param.default is inspect.Parameter.empty:
- raise TypeError(f'Required parameter {key} is empty.')
+ raise TypeError(f"Required parameter {key} is empty.")
data = param.default
out[key] = data
return out
+
def func_header_repr(func: Callable, keep_annotation: bool = True) -> str:
source = inspect.getsource(func)
signature = inspect.signature(func)
if keep_annotation:
- return ''.join(re.split(r'(\).*?:.*?\n)', source,
- maxsplit=1)[:2]).strip()
+ return "".join(re.split(r"(\).*?:.*?\n)", source, maxsplit=1)[:2]).strip()
- params_repr = ['self']
+ params_repr = ["self"]
for param in signature.parameters.values():
params_repr.append(param.name)
if param.default is not inspect.Parameter.empty:
- params_repr[-1] += f'={param.default}'
+ params_repr[-1] += f"={param.default}"
return f'def {func.__name__}({", ".join(params_repr)}):'
+
def func_body_repr(func: Callable, keep_annotation: bool = True) -> str:
source = inspect.getsource(func)
- body_repr = re.split(r'\).*?:.*?\n', source, maxsplit=1)[1]
+ body_repr = re.split(r"\).*?:.*?\n", source, maxsplit=1)[1]
if not keep_annotation:
- body_repr = re.sub(r'\s*# type:.*\n', '', body_repr)
- return body_repr
\ No newline at end of file
+ body_repr = re.sub(r"\s*# type:.*\n", "", body_repr)
+ return body_repr
diff --git a/jointContribution/graphGalerkin/utils/linear.py b/jointContribution/graphGalerkin/utils/linear.py
index b74f953c3e..861008ebf2 100644
--- a/jointContribution/graphGalerkin/utils/linear.py
+++ b/jointContribution/graphGalerkin/utils/linear.py
@@ -1,16 +1,15 @@
-from typing import Optional
-from collections import OrderedDict
-
import copy
import math
+from collections import OrderedDict
+from typing import Optional
+import init as inits
import paddle
-from paddle import nn
-from paddle import Tensor
import paddle.nn.functional as F
+from paddle import Tensor
+from paddle import nn
from paddle.nn.initializer import Uniform
-import init as inits
class Linear(nn.Layer):
r"""Applies a linear tranformation to the incoming data
@@ -38,9 +37,15 @@ class Linear(nn.Layer):
If set to :obj:`None`, will match default bias initialization of
:class:`torch.nn.Linear`. (default: :obj:`None`)
"""
- def __init__(self, in_channels: int, out_channels: int, bias: bool = True,
- weight_initializer: Optional[str] = None,
- bias_initializer: Optional[str] = None):
+
+ def __init__(
+ self,
+ in_channels: int,
+ out_channels: int,
+ bias: bool = True,
+ weight_initializer: Optional[str] = None,
+ bias_initializer: Optional[str] = None,
+ ):
super().__init__()
self.in_channels = in_channels
self.out_channels = out_channels
@@ -49,26 +54,32 @@ def __init__(self, in_channels: int, out_channels: int, bias: bool = True,
self._parameters = OrderedDict()
if in_channels > 0:
- self.weight = paddle.create_parameter(shape=[in_channels, out_channels], dtype=paddle.float32)
+ self.weight = paddle.create_parameter(
+ shape=[in_channels, out_channels], dtype=paddle.float32
+ )
else:
self.weight = nn.parameter.UninitializedParameter()
- self._hook = self.register_forward_pre_hook(
- self.initialize_parameters)
+ self._hook = self.register_forward_pre_hook(self.initialize_parameters)
if bias:
- self.bias = paddle.create_parameter(shape=[out_channels], dtype=paddle.float32)
+ self.bias = paddle.create_parameter(
+ shape=[out_channels], dtype=paddle.float32
+ )
else:
- self.register_parameter('bias', None)
+ self.register_parameter("bias", None)
- self._load_hook = self.register_state_dict_hook(
- self._lazy_load_hook)
+ self._load_hook = self.register_state_dict_hook(self._lazy_load_hook)
self.reset_parameters()
def __deepcopy__(self, memo):
- out = Linear(self.in_channels, self.out_channels, self.bias
- is not None, self.weight_initializer,
- self.bias_initializer)
+ out = Linear(
+ self.in_channels,
+ self.out_channels,
+ self.bias is not None,
+ self.weight_initializer,
+ self.bias_initializer,
+ )
if self.in_channels > 0:
out.weight = copy.deepcopy(self.weight, memo)
if self.bias is not None:
@@ -77,35 +88,41 @@ def __deepcopy__(self, memo):
def reset_parameters(self):
# if isinstance(self.weight, nn.parameter.UninitializedParameter):
- if self.in_channels<=0:
+ if self.in_channels <= 0:
pass
- elif self.weight_initializer == 'glorot':
+ elif self.weight_initializer == "glorot":
inits.glorot(self.weight)
- elif self.weight_initializer == 'uniform':
+ elif self.weight_initializer == "uniform":
bound = 1.0 / math.sqrt(self.weight.size(-1))
- self.weight = paddle.create_parameter(shape=self.weight.shape, dtype=paddle.float32, attr=paddle.ParamAttr(initializer=Uniform(-bound, bound)))
- elif self.weight_initializer == 'kaiming_uniform':
- inits.kaiming_uniform(self.weight, fan=self.in_channels,
- a=math.sqrt(5))
+ self.weight = paddle.create_parameter(
+ shape=self.weight.shape,
+ dtype=paddle.float32,
+ attr=paddle.ParamAttr(initializer=Uniform(-bound, bound)),
+ )
+ elif self.weight_initializer == "kaiming_uniform":
+ inits.kaiming_uniform(self.weight, fan=self.in_channels, a=math.sqrt(5))
elif self.weight_initializer is None:
- inits.kaiming_uniform(self.weight, fan=self.in_channels,
- a=math.sqrt(5))
+ inits.kaiming_uniform(self.weight, fan=self.in_channels, a=math.sqrt(5))
else:
- raise RuntimeError(f"Linear layer weight initializer "
- f"'{self.weight_initializer}' is not supported")
+ raise RuntimeError(
+ f"Linear layer weight initializer "
+ f"'{self.weight_initializer}' is not supported"
+ )
# if isinstance(self.weight, nn.parameter.UninitializedParameter):
- if self.in_channels<=0:
+ if self.in_channels <= 0:
pass
elif self.bias is None:
pass
- elif self.bias_initializer == 'zeros':
+ elif self.bias_initializer == "zeros":
inits.zeros(self.bias)
elif self.bias_initializer is None:
inits.uniform(self.in_channels, self.bias)
else:
- raise RuntimeError(f"Linear layer bias initializer "
- f"'{self.bias_initializer}' is not supported")
+ raise RuntimeError(
+ f"Linear layer bias initializer "
+ f"'{self.bias_initializer}' is not supported"
+ )
def forward(self, x: Tensor) -> Tensor:
""""""
@@ -118,41 +135,49 @@ def initialize_parameters(self, module, input):
self.weight.materialize((self.out_channels, self.in_channels))
self.reset_parameters()
self._hook.remove()
- delattr(self, '_hook')
+ delattr(self, "_hook")
def _save_to_state_dict(self, destination, prefix, keep_vars):
if isinstance(self.weight, nn.parameter.UninitializedParameter):
- destination[prefix + 'weight'] = self.weight
+ destination[prefix + "weight"] = self.weight
else:
- destination[prefix + 'weight'] = self.weight.detach()
+ destination[prefix + "weight"] = self.weight.detach()
if self.bias is not None:
- destination[prefix + 'bias'] = self.bias.detach()
-
- def _lazy_load_hook(self, state_dict, prefix, local_metadata, strict,
- missing_keys, unexpected_keys, error_msgs):
-
- weight = state_dict[prefix + 'weight']
+ destination[prefix + "bias"] = self.bias.detach()
+
+ def _lazy_load_hook(
+ self,
+ state_dict,
+ prefix,
+ local_metadata,
+ strict,
+ missing_keys,
+ unexpected_keys,
+ error_msgs,
+ ):
+
+ weight = state_dict[prefix + "weight"]
if isinstance(weight, nn.parameter.UninitializedParameter):
self.in_channels = -1
self.weight = nn.parameter.UninitializedParameter()
- if not hasattr(self, '_hook'):
- self._hook = self.register_forward_pre_hook(
- self.initialize_parameters)
+ if not hasattr(self, "_hook"):
+ self._hook = self.register_forward_pre_hook(self.initialize_parameters)
elif isinstance(self.weight, nn.parameter.UninitializedParameter):
self.in_channels = weight.size(-1)
self.weight.materialize((self.out_channels, self.in_channels))
- if hasattr(self, '_hook'):
+ if hasattr(self, "_hook"):
self._hook.remove()
- delattr(self, '_hook')
+ delattr(self, "_hook")
def __repr__(self) -> str:
- return (f'{self.__class__.__name__}({self.in_channels}, '
- f'{self.out_channels}, bias={self.bias is not None})')
-
+ return (
+ f"{self.__class__.__name__}({self.in_channels}, "
+ f"{self.out_channels}, bias={self.bias is not None})"
+ )
+
def register_parameter(self, name: str, param) -> None:
if param is None:
self._parameters[name] = None
else:
self._parameters[name] = param
-
diff --git a/jointContribution/graphGalerkin/utils/message_passing.py b/jointContribution/graphGalerkin/utils/message_passing.py
index 98ef276f2b..e36966a058 100644
--- a/jointContribution/graphGalerkin/utils/message_passing.py
+++ b/jointContribution/graphGalerkin/utils/message_passing.py
@@ -1,17 +1,19 @@
-from typing import List, Optional, Set, Tuple
-
-from uuid import uuid1
-from inspect import Parameter
from collections import OrderedDict
+from inspect import Parameter
+from typing import List
+from typing import Optional
+from typing import Set
+from typing import Tuple
import paddle
-from paddle import Tensor
-
from inspector import Inspector
-from scatter import scatter
+from paddle import Tensor
from paddle.nn import Layer
+from scatter import scatter
Size = Optional[Tuple[int, int]]
+
+
class MessagePassing(Layer):
r"""Base class for creating message passing layers of the form
@@ -55,21 +57,33 @@ class MessagePassing(Layer):
"""
special_args: Set[str] = {
- 'edge_index', 'adj_t', 'edge_index_i', 'edge_index_j', 'size',
- 'size_i', 'size_j', 'ptr', 'index', 'dim_size'
+ "edge_index",
+ "adj_t",
+ "edge_index_i",
+ "edge_index_j",
+ "size",
+ "size_i",
+ "size_j",
+ "ptr",
+ "index",
+ "dim_size",
}
- def __init__(self, aggr: Optional[str] = "add",
- flow: str = "source_to_target", node_dim: int = -2,
- decomposed_layers: int = 1):
+ def __init__(
+ self,
+ aggr: Optional[str] = "add",
+ flow: str = "source_to_target",
+ node_dim: int = -2,
+ decomposed_layers: int = 1,
+ ):
super().__init__()
self.aggr = aggr
- assert self.aggr in ['add', 'mean', 'max', None]
+ assert self.aggr in ["add", "mean", "max", None]
self.flow = flow
- assert self.flow in ['source_to_target', 'target_to_source']
+ assert self.flow in ["source_to_target", "target_to_source"]
self.node_dim = node_dim
self.decomposed_layers = decomposed_layers
@@ -82,14 +96,17 @@ def __init__(self, aggr: Optional[str] = "add",
self.inspector.inspect(self.message_and_aggregate, pop_first=True)
self.__user_args__ = self.inspector.keys(
- ['message', 'aggregate', 'update']).difference(self.special_args)
+ ["message", "aggregate", "update"]
+ ).difference(self.special_args)
self.__fused_user_args__ = self.inspector.keys(
- ['message_and_aggregate', 'update']).difference(self.special_args)
- self.__edge_user_args__ = self.inspector.keys(
- ['edge_update']).difference(self.special_args)
+ ["message_and_aggregate", "update"]
+ ).difference(self.special_args)
+ self.__edge_user_args__ = self.inspector.keys(["edge_update"]).difference(
+ self.special_args
+ )
# Support for "fused" message passing.
- self.fuse = self.inspector.implements('message_and_aggregate')
+ self.fuse = self.inspector.implements("message_and_aggregate")
# Support for GNNExplainer.
self.__explain__ = False
@@ -114,24 +131,27 @@ def __set_size__(self, size: List[Optional[int]], dim: int, src: Tensor):
size[dim] = src.shape[self.node_dim]
elif the_size != src.shape[self.node_dim]:
raise ValueError(
- (f'Encountered tensor with size {src.size(self.node_dim)} in '
- f'dimension {self.node_dim}, but expected size {the_size}.'))
-
+ (
+ f"Encountered tensor with size {src.size(self.node_dim)} in "
+ f"dimension {self.node_dim}, but expected size {the_size}."
+ )
+ )
+
def __lift__(self, src, edge_index, dim):
if isinstance(edge_index, Tensor):
index = edge_index[dim]
return src.index_select(index, self.node_dim)
raise ValueError
-
+
def __collect__(self, args, edge_index, size, kwargs):
- i, j = (1, 0) if self.flow == 'source_to_target' else (0, 1)
+ i, j = (1, 0) if self.flow == "source_to_target" else (0, 1)
out = {}
for arg in args:
- if arg[-2:] not in ['_i', '_j']:
+ if arg[-2:] not in ["_i", "_j"]:
out[arg] = kwargs.get(arg, Parameter.empty)
else:
- dim = 0 if arg[-2:] == '_j' else 1
+ dim = 0 if arg[-2:] == "_j" else 1
data = kwargs.get(arg[:-2], Parameter.empty)
if isinstance(data, (tuple, list)):
@@ -142,26 +162,25 @@ def __collect__(self, args, edge_index, size, kwargs):
if isinstance(data, Tensor):
self.__set_size__(size, dim, data)
- data = self.__lift__(data, edge_index,
- j if arg[-2:] == '_j' else i)
+ data = self.__lift__(data, edge_index, j if arg[-2:] == "_j" else i)
out[arg] = data
if isinstance(edge_index, Tensor):
- out['adj_t'] = None
- out['edge_index'] = edge_index
- out['edge_index_i'] = edge_index[i]
- out['edge_index_j'] = edge_index[j]
- out['ptr'] = None
-
- out['index'] = out['edge_index_i']
- out['size'] = size
- out['size_i'] = size[1] or size[0]
- out['size_j'] = size[0] or size[1]
- out['dim_size'] = out['size_i']
+ out["adj_t"] = None
+ out["edge_index"] = edge_index
+ out["edge_index_i"] = edge_index[i]
+ out["edge_index_j"] = edge_index[j]
+ out["ptr"] = None
+
+ out["index"] = out["edge_index_i"]
+ out["size"] = size
+ out["size_i"] = size[1] or size[0]
+ out["size_j"] = size[0] or size[1]
+ out["dim_size"] = out["size_i"]
return out
-
+
def __check_input__(self, edge_index, size):
the_size: List[Optional[int]] = [None, None]
@@ -175,9 +194,12 @@ def __check_input__(self, edge_index, size):
return the_size
raise ValueError(
- ('`MessagePassing.propagate` only supports `int` of '
- 'shape `[2, num_messages]` for '
- 'argument `edge_index`.'))
+ (
+ "`MessagePassing.propagate` only supports `int` of "
+ "shape `[2, num_messages]` for "
+ "argument `edge_index`."
+ )
+ )
def propagate(self, edge_index: Tensor, size: Size = None, **kwargs):
r"""The initial call to start propagating messages.
@@ -216,10 +238,9 @@ def propagate(self, edge_index: Tensor, size: Size = None, **kwargs):
if isinstance(edge_index, Tensor) or not self.fuse:
if decomposed_layers > 1:
user_args = self.__user_args__
- decomp_args = {a[:-2] for a in user_args if a[-2:] == '_j'}
+ decomp_args = {a[:-2] for a in user_args if a[-2:] == "_j"}
decomp_kwargs = {
- a: kwargs[a].chunk(decomposed_layers, -1)
- for a in decomp_args
+ a: kwargs[a].chunk(decomposed_layers, -1) for a in decomp_args
}
decomp_out = []
@@ -228,17 +249,18 @@ def propagate(self, edge_index: Tensor, size: Size = None, **kwargs):
for arg in decomp_args:
kwargs[arg] = decomp_kwargs[arg][i]
- coll_dict = self.__collect__(self.__user_args__, edge_index,
- size, kwargs)
+ coll_dict = self.__collect__(
+ self.__user_args__, edge_index, size, kwargs
+ )
- msg_kwargs = self.inspector.distribute('message', coll_dict)
+ msg_kwargs = self.inspector.distribute("message", coll_dict)
for hook in self._message_forward_pre_hooks.values():
- res = hook(self, (msg_kwargs, ))
+ res = hook(self, (msg_kwargs,))
if res is not None:
msg_kwargs = res[0] if isinstance(res, tuple) else res
out = self.message(**msg_kwargs)
for hook in self._message_forward_hooks.values():
- res = hook(self, (msg_kwargs, ), out)
+ res = hook(self, (msg_kwargs,), out)
if res is not None:
out = res
@@ -256,19 +278,19 @@ def propagate(self, edge_index: Tensor, size: Size = None, **kwargs):
assert out.size(self.node_dim) == edge_mask.size(0)
out = out * edge_mask.view([-1] + [1] * (out.dim() - 1))
- aggr_kwargs = self.inspector.distribute('aggregate', coll_dict)
+ aggr_kwargs = self.inspector.distribute("aggregate", coll_dict)
for hook in self._aggregate_forward_pre_hooks.values():
- res = hook(self, (aggr_kwargs, ))
+ res = hook(self, (aggr_kwargs,))
if res is not None:
aggr_kwargs = res[0] if isinstance(res, tuple) else res
out = self.aggregate(out, **aggr_kwargs)
for hook in self._aggregate_forward_hooks.values():
- res = hook(self, (aggr_kwargs, ), out)
+ res = hook(self, (aggr_kwargs,), out)
if res is not None:
out = res
- update_kwargs = self.inspector.distribute('update', coll_dict)
+ update_kwargs = self.inspector.distribute("update", coll_dict)
out = self.update(out, **update_kwargs)
if decomposed_layers > 1:
@@ -283,7 +305,7 @@ def propagate(self, edge_index: Tensor, size: Size = None, **kwargs):
out = res
return out
-
+
def message(self, x_j: Tensor) -> Tensor:
r"""Constructs messages from node :math:`j` to node :math:`i`
in analogy to :math:`\phi_{\mathbf{\Theta}}` for each edge in
@@ -295,10 +317,14 @@ def message(self, x_j: Tensor) -> Tensor:
:obj:`_j` to the variable name, *.e.g.* :obj:`x_i` and :obj:`x_j`.
"""
return x_j
-
- def aggregate(self, inputs: Tensor, index: Tensor,
- ptr: Optional[Tensor] = None,
- dim_size: Optional[int] = None) -> Tensor:
+
+ def aggregate(
+ self,
+ inputs: Tensor,
+ index: Tensor,
+ ptr: Optional[Tensor] = None,
+ dim_size: Optional[int] = None,
+ ) -> Tensor:
r"""Aggregates messages from neighbors as
:math:`\square_{j \in \mathcal{N}(i)}`.
@@ -309,9 +335,10 @@ def aggregate(self, inputs: Tensor, index: Tensor,
that support "add", "mean" and "max" operations as specified in
:meth:`__init__` by the :obj:`aggr` argument.
"""
- return scatter(inputs, index, dim=self.node_dim, dim_size=dim_size,
- reduce=self.aggr)
-
+ return scatter(
+ inputs, index, dim=self.node_dim, dim_size=dim_size, reduce=self.aggr
+ )
+
def update(self, inputs: Tensor) -> Tensor:
r"""Updates node embeddings in analogy to
:math:`\gamma_{\mathbf{\Theta}}` for each node
@@ -320,7 +347,7 @@ def update(self, inputs: Tensor) -> Tensor:
which was initially passed to :meth:`propagate`.
"""
return inputs
-
+
def edge_update(self) -> Tensor:
r"""Computes or updates features for each edge in the graph.
This function can take any argument as input which was initially passed
@@ -330,6 +357,7 @@ def edge_update(self) -> Tensor:
:obj:`_j` to the variable name, *.e.g.* :obj:`x_i` and :obj:`x_j`.
"""
raise NotImplementedError
+
def message_and_aggregate(self, adj_t) -> Tensor:
r"""Fuses computations of :func:`message` and :func:`aggregate` into a
single function.
@@ -338,4 +366,4 @@ def message_and_aggregate(self, adj_t) -> Tensor:
This function will only gets called in case it is implemented and
propagation takes place based on a :obj:`torch_sparse.SparseTensor`.
"""
- raise NotImplementedError
\ No newline at end of file
+ raise NotImplementedError
diff --git a/jointContribution/graphGalerkin/utils/scatter.py b/jointContribution/graphGalerkin/utils/scatter.py
index eebea65be9..edcb82f83c 100644
--- a/jointContribution/graphGalerkin/utils/scatter.py
+++ b/jointContribution/graphGalerkin/utils/scatter.py
@@ -1,7 +1,8 @@
-from typing import Optional, Tuple
+from typing import Optional
import paddle
+
def broadcast(src: paddle.Tensor, other: paddle.Tensor, dim: int):
if dim < 0:
dim = other.dim() + dim
@@ -13,21 +14,29 @@ def broadcast(src: paddle.Tensor, other: paddle.Tensor, dim: int):
src = src.expand_as(other)
return src
+
def scatter_add_(dim, index, src, x):
- if x.dim()==1:
- output = paddle.scatter_nd_add(x.unsqueeze(-1), index.unsqueeze(-1), src.unsqueeze(-1)).squeeze(-1)
+ if x.dim() == 1:
+ output = paddle.scatter_nd_add(
+ x.unsqueeze(-1), index.unsqueeze(-1), src.unsqueeze(-1)
+ ).squeeze(-1)
else:
i, j = index.shape
- grid_x , grid_y = paddle.meshgrid(paddle.arange(i), paddle.arange(j))
+ grid_x, grid_y = paddle.meshgrid(paddle.arange(i), paddle.arange(j))
index = paddle.stack([index.flatten(), grid_y.flatten()], axis=1)
updates_index = paddle.stack([grid_x.flatten(), grid_y.flatten()], axis=1)
updates = paddle.gather_nd(src, index=updates_index)
output = paddle.scatter_nd_add(x, index, updates)
return output
-def scatter_sum(src: paddle.Tensor, index: paddle.Tensor, dim: int = -1,
- out: Optional[paddle.Tensor] = None,
- dim_size: Optional[int] = None) -> paddle.Tensor:
+
+def scatter_sum(
+ src: paddle.Tensor,
+ index: paddle.Tensor,
+ dim: int = -1,
+ out: Optional[paddle.Tensor] = None,
+ dim_size: Optional[int] = None,
+) -> paddle.Tensor:
index = broadcast(index, src, dim)
if out is None:
size = list(src.shape)
@@ -42,14 +51,24 @@ def scatter_sum(src: paddle.Tensor, index: paddle.Tensor, dim: int = -1,
else:
return scatter_add_(0, index, src, out)
-def scatter_add(src: paddle.Tensor, index: paddle.Tensor, dim: int = -1,
- out: Optional[paddle.Tensor] = None,
- dim_size: Optional[int] = None) -> paddle.Tensor:
+
+def scatter_add(
+ src: paddle.Tensor,
+ index: paddle.Tensor,
+ dim: int = -1,
+ out: Optional[paddle.Tensor] = None,
+ dim_size: Optional[int] = None,
+) -> paddle.Tensor:
return scatter_sum(src, index, dim, out, dim_size)
-def scatter_mean(src: paddle.Tensor, index: paddle.Tensor, dim: int = -1,
- out: Optional[paddle.Tensor] = None,
- dim_size: Optional[int] = None) -> paddle.Tensor:
+
+def scatter_mean(
+ src: paddle.Tensor,
+ index: paddle.Tensor,
+ dim: int = -1,
+ out: Optional[paddle.Tensor] = None,
+ dim_size: Optional[int] = None,
+) -> paddle.Tensor:
out = scatter_sum(src, index, dim, out, dim_size)
dim_size = out.size(dim)
@@ -70,9 +89,15 @@ def scatter_mean(src: paddle.Tensor, index: paddle.Tensor, dim: int = -1,
out.floor_divide_(count)
return out
-def scatter(src: paddle.Tensor, index: paddle.Tensor, dim: int = -1,
- out: Optional[paddle.Tensor] = None, dim_size: Optional[int] = None,
- reduce: str = "sum") -> paddle.Tensor:
+
+def scatter(
+ src: paddle.Tensor,
+ index: paddle.Tensor,
+ dim: int = -1,
+ out: Optional[paddle.Tensor] = None,
+ dim_size: Optional[int] = None,
+ reduce: str = "sum",
+) -> paddle.Tensor:
r"""
Reduces all values from the :attr:`src` tensor into :attr:`out` at the
indices specified in the :attr:`index` tensor along a given axis
@@ -121,9 +146,9 @@ def scatter(src: paddle.Tensor, index: paddle.Tensor, dim: int = -1,
:rtype: :class:`Tensor`
"""
- if reduce == 'sum' or reduce == 'add':
+ if reduce == "sum" or reduce == "add":
return scatter_sum(src, index, dim, out, dim_size)
- elif reduce == 'mean':
+ elif reduce == "mean":
return scatter_mean(src, index, dim, out, dim_size)
else:
raise ValueError
diff --git a/jointContribution/graphGalerkin/utils/utils.py b/jointContribution/graphGalerkin/utils/utils.py
index c495408bd6..2764f02771 100644
--- a/jointContribution/graphGalerkin/utils/utils.py
+++ b/jointContribution/graphGalerkin/utils/utils.py
@@ -1,22 +1,29 @@
-from paddle import Tensor
-from typing import Optional, Tuple, Union
+from typing import Optional
+from typing import Tuple
+from typing import Union
+
import paddle
+from paddle import Tensor
+from scatter import scatter
+from scatter import scatter_add
-from scatter import scatter, scatter_add
OptTensor = Optional[Tensor]
import pgl
-class Data():
+
+class Data:
def __init__(self, x, y, edge_index):
self.y = y
self.x = x
self.edge_index = edge_index
+
def __call__(self):
- return pgl.Graph(edges=self.edge_index,
- num_nodes=self.x.shape[0],
- node_feat=self.x)
-
+ return pgl.Graph(
+ edges=self.edge_index, num_nodes=self.x.shape[0], node_feat=self.x
+ )
+
+
def maybe_num_nodes(edge_index, num_nodes=None):
if num_nodes is not None:
return num_nodes
@@ -25,8 +32,10 @@ def maybe_num_nodes(edge_index, num_nodes=None):
else:
return max(edge_index.size(0), edge_index.size(1))
-def remove_self_loops(edge_index: Tensor,
- edge_attr: OptTensor = None) -> Tuple[Tensor, OptTensor]:
+
+def remove_self_loops(
+ edge_index: Tensor, edge_attr: OptTensor = None
+) -> Tuple[Tensor, OptTensor]:
r"""Removes every self-loop in the graph given by :attr:`edge_index`, so
that :math:`(i,i) \not\in \mathcal{E}` for every :math:`i \in \mathcal{V}`.
@@ -39,16 +48,19 @@ def remove_self_loops(edge_index: Tensor,
"""
mask = edge_index[0] != edge_index[1]
for _ in range(edge_index.dim()):
- edge_index[_] = paddle.masked_select(edge_index[_], mask)
+ edge_index[_] = paddle.masked_select(edge_index[_], mask)
if edge_attr is None:
return edge_index, None
else:
return edge_index, edge_attr[mask]
+
def add_self_loops(
- edge_index: Tensor, edge_attr: OptTensor = None,
- fill_value: Union[float, Tensor, str] = None,
- num_nodes: Optional[int] = None) -> Tuple[Tensor, OptTensor]:
+ edge_index: Tensor,
+ edge_attr: OptTensor = None,
+ fill_value: Union[float, Tensor, str] = None,
+ num_nodes: Optional[int] = None,
+) -> Tuple[Tensor, OptTensor]:
r"""Adds a self-loop :math:`(i,i) \in \mathcal{E}` to every node
:math:`i \in \mathcal{V}` in the graph given by :attr:`edge_index`.
In case the graph is weighted or has multi-dimensional edge features
@@ -78,10 +90,10 @@ def add_self_loops(
loop_index = paddle.tile(loop_index.unsqueeze(0), repeat_times=[2, 1])
if edge_attr is not None:
if fill_value is None:
- loop_attr = edge_attr.new_full((N, ) + edge_attr.size()[1:], 1.)
+ loop_attr = edge_attr.new_full((N,) + edge_attr.size()[1:], 1.0)
elif isinstance(fill_value, (int, float)):
- loop_attr = paddle.full((N, ), fill_value, dtype=edge_attr.dtype)
+ loop_attr = paddle.full((N,), fill_value, dtype=edge_attr.dtype)
elif isinstance(fill_value, Tensor):
loop_attr = fill_value.to(edge_attr.device, edge_attr.dtype)
if edge_attr.dim() != loop_attr.dim():
@@ -90,21 +102,26 @@ def add_self_loops(
loop_attr = loop_attr.repeat(*sizes)
elif isinstance(fill_value, str):
- loop_attr = scatter(edge_attr, edge_index[1], dim=0, dim_size=N,
- reduce=fill_value)
+ loop_attr = scatter(
+ edge_attr, edge_index[1], dim=0, dim_size=N, reduce=fill_value
+ )
else:
raise AttributeError("No valid 'fill_value' provided")
-
+
edge_attr = paddle.concat([edge_attr, loop_attr], axis=0)
edge_index = paddle.concat([edge_index, loop_index], axis=1)
return edge_index, edge_attr
-
-def get_laplacian(edge_index, edge_weight: Optional[paddle.Tensor] = None,
- normalization: Optional[str] = None,
- dtype: Optional[int] = None,
- num_nodes: Optional[int] = None):
- r""" Computes the graph Laplacian of the graph given by :obj:`edge_index`
+
+
+def get_laplacian(
+ edge_index,
+ edge_weight: Optional[paddle.Tensor] = None,
+ normalization: Optional[str] = None,
+ dtype: Optional[int] = None,
+ num_nodes: Optional[int] = None,
+):
+ r"""Computes the graph Laplacian of the graph given by :obj:`edge_index`
and optional :obj:`edge_weight`.
Args:
@@ -130,7 +147,7 @@ def get_laplacian(edge_index, edge_weight: Optional[paddle.Tensor] = None,
"""
if normalization is not None:
- assert normalization in ['sym', 'rw'] # 'Invalid normalization'
+ assert normalization in ["sym", "rw"] # 'Invalid normalization'
edge_index, edge_weight = remove_self_loops(edge_index, edge_weight)
@@ -144,38 +161,44 @@ def get_laplacian(edge_index, edge_weight: Optional[paddle.Tensor] = None,
# L = D - A.
edge_index, _ = add_self_loops(edge_index, num_nodes=num_nodes)
edge_weight = paddle.concat([-edge_weight, deg], dim=0)
- elif normalization == 'sym':
+ elif normalization == "sym":
# Compute A_norm = -D^{-1/2} A D^{-1/2}.
deg_inv_sqrt = deg.pow(-0.5)
- deg_inv_sqrt = masked_fill(deg_inv_sqrt,deg_inv_sqrt == float('inf'), 0)
+ deg_inv_sqrt = masked_fill(deg_inv_sqrt, deg_inv_sqrt == float("inf"), 0)
edge_weight = deg_inv_sqrt[row] * edge_weight * deg_inv_sqrt[col]
# L = I - A_norm.
- edge_index, tmp = add_self_loops(edge_index, -edge_weight,
- fill_value=1., num_nodes=num_nodes)
+ edge_index, tmp = add_self_loops(
+ edge_index, -edge_weight, fill_value=1.0, num_nodes=num_nodes
+ )
assert tmp is not None
edge_weight = tmp
else:
# Compute A_norm = -D^{-1} A.
deg_inv = 1.0 / deg
- deg_inv.masked_fill_(deg_inv == float('inf'), 0)
+ deg_inv.masked_fill_(deg_inv == float("inf"), 0)
edge_weight = deg_inv[row] * edge_weight
# L = I - A_norm.
- edge_index, tmp = add_self_loops(edge_index, -edge_weight,
- fill_value=1., num_nodes=num_nodes)
+ edge_index, tmp = add_self_loops(
+ edge_index, -edge_weight, fill_value=1.0, num_nodes=num_nodes
+ )
assert tmp is not None
edge_weight = tmp
return edge_index, edge_weight
+
def masked_fill(x, mask, value):
y = paddle.full(x.shape, value, x.dtype)
return paddle.where(mask, y, x)
+
def add_remaining_self_loops(
- edge_index: Tensor, edge_attr: OptTensor = None,
- fill_value: Union[float, Tensor, str] = None,
- num_nodes: Optional[int] = None) -> Tuple[Tensor, OptTensor]:
+ edge_index: Tensor,
+ edge_attr: OptTensor = None,
+ fill_value: Union[float, Tensor, str] = None,
+ num_nodes: Optional[int] = None,
+) -> Tuple[Tensor, OptTensor]:
r"""Adds remaining self-loop :math:`(i,i) \in \mathcal{E}` to every node
:math:`i \in \mathcal{V}` in the graph given by :attr:`edge_index`.
In case the graph is weighted or has multi-dimensional edge features
@@ -200,17 +223,17 @@ def add_remaining_self_loops(
:rtype: (:class:`LongTensor`, :class:`Tensor`)
"""
N = maybe_num_nodes(edge_index, num_nodes)
- mask = edge_index[0] != edge_index[1]
+ edge_index[0] != edge_index[1]
loop_index = paddle.arange(0, N, dtype=paddle.int32)
loop_index = paddle.tile(loop_index.unsqueeze(0), repeat_times=[2, 1])
if edge_attr is not None:
if fill_value is None:
- loop_attr = edge_attr.new_full((N, ) + edge_attr.size()[1:], 1.)
+ loop_attr = edge_attr.new_full((N,) + edge_attr.size()[1:], 1.0)
elif isinstance(fill_value, (int, float)):
- loop_attr = paddle.full((N, ), fill_value, dtype=edge_attr.dtype)
+ loop_attr = paddle.full((N,), fill_value, dtype=edge_attr.dtype)
elif isinstance(fill_value, Tensor):
loop_attr = fill_value.to(edge_attr.device, edge_attr.dtype)
if edge_attr.dim() != loop_attr.dim():
@@ -219,18 +242,18 @@ def add_remaining_self_loops(
loop_attr = loop_attr.repeat(*sizes)
elif isinstance(fill_value, str):
- loop_attr = scatter(edge_attr, edge_index[1], dim=0, dim_size=N,
- reduce=fill_value)
+ loop_attr = scatter(
+ edge_attr, edge_index[1], dim=0, dim_size=N, reduce=fill_value
+ )
else:
raise AttributeError("No valid 'fill_value' provided")
- inv_mask = ~mask
-
edge_attr = paddle.concat([edge_attr, loop_attr], axis=0)
edge_index = paddle.concat([edge_index, loop_index], axis=1)
return edge_index, edge_attr
+
def expand_left(src: paddle.Tensor, dim: int, dims: int) -> paddle.Tensor:
for _ in range(dims + dim if dim < 0 else dim):
src = src.unsqueeze(0)
- return src
\ No newline at end of file
+ return src
diff --git a/mkdocs.yml b/mkdocs.yml
index 2e70c8092b..bfc899dfed 100644
--- a/mkdocs.yml
+++ b/mkdocs.yml
@@ -100,6 +100,7 @@ nav:
- FourCastNet: zh/examples/fourcastnet.md
- NowcastNet: zh/examples/nowcastnet.md
- DGMR: zh/examples/dgmr.md
+ - STAFNet: zh/examples/stafnet.md
- EarthFormer: zh/examples/earthformer.md
- GraphCast: zh/examples/graphcast.md
- GenCast: zh/examples/gencast.md
diff --git a/ppsci/arch/__init__.py b/ppsci/arch/__init__.py
index ec3f63597c..d0d99db16b 100644
--- a/ppsci/arch/__init__.py
+++ b/ppsci/arch/__init__.py
@@ -62,6 +62,7 @@
from ppsci.arch.regdgcnn import RegDGCNN # isort:skip
from ppsci.arch.regpointnet import RegPointNet # isort:skip
from ppsci.arch.ifm_mlp import IFMMLP # isort:skip
+from ppsci.arch.stafnet import STAFNet # isort:skip
__all__ = [
"MoFlowNet",
@@ -113,6 +114,7 @@
"RegDGCNN",
"RegPointNet",
"IFMMLP",
+ "STAFNet",
]
diff --git a/ppsci/arch/stafnet.py b/ppsci/arch/stafnet.py
new file mode 100644
index 0000000000..293a3416a5
--- /dev/null
+++ b/ppsci/arch/stafnet.py
@@ -0,0 +1,747 @@
+import math
+from math import sqrt
+from typing import Tuple
+
+import numpy as np
+import paddle
+
+from ppsci.arch import base
+
+try:
+ from pgl.nn.conv import GATv2Conv
+except ModuleNotFoundError:
+ pass
+
+
+class Inception_Block_V1(paddle.nn.Layer):
+ def __init__(self, in_channels, out_channels, num_kernels=6, init_weight=True):
+ super(Inception_Block_V1, self).__init__()
+ self.in_channels = in_channels
+ self.out_channels = out_channels
+ self.num_kernels = num_kernels
+ kernels = []
+ for i in range(self.num_kernels):
+ kernels.append(
+ paddle.nn.Conv2D(
+ in_channels=in_channels,
+ out_channels=out_channels,
+ kernel_size=2 * i + 1,
+ padding=i,
+ )
+ )
+ self.kernels = paddle.nn.LayerList(sublayers=kernels)
+ if init_weight:
+ self._initialize_weights()
+
+ def _initialize_weights(self):
+ for m in self.sublayers():
+ if isinstance(m, paddle.nn.Conv2D):
+
+ init_kaimingNormal = paddle.nn.initializer.KaimingNormal(
+ fan_in=None, negative_slope=0.0, nonlinearity="relu"
+ )
+ init_kaimingNormal(m.weight)
+
+ if m.bias is not None:
+ init_Constant = paddle.nn.initializer.Constant(value=0)
+ init_Constant(m.bias)
+
+ def forward(self, x):
+ res_list = []
+ for i in range(self.num_kernels):
+ res_list.append(self.kernels[i](x))
+ res = paddle.stack(x=res_list, axis=-1).mean(axis=-1)
+ return res
+
+
+class AttentionLayer(paddle.nn.Layer):
+ def __init__(self, attention, d_model, n_heads, d_keys=None, d_values=None):
+ super(AttentionLayer, self).__init__()
+ d_keys = d_keys or d_model // n_heads
+ d_values = d_values or d_model // n_heads
+ self.inner_attention = attention
+ self.query_projection = paddle.nn.Linear(
+ in_features=d_model, out_features=d_keys * n_heads
+ )
+ self.key_projection = paddle.nn.Linear(
+ in_features=d_model, out_features=d_keys * n_heads
+ )
+ self.value_projection = paddle.nn.Linear(
+ in_features=d_model, out_features=d_values * n_heads
+ )
+ self.out_projection = paddle.nn.Linear(
+ in_features=d_values * n_heads, out_features=d_model
+ )
+ self.n_heads = n_heads
+
+ def forward(self, queries, keys, values, attn_mask):
+ B, L, _ = tuple(queries.shape)
+ _, S, _ = tuple(keys.shape)
+ H = self.n_heads
+ queries = self.query_projection(queries).reshape((B, L, H, -1))
+ keys = self.key_projection(keys).reshape((B, S, H, -1))
+ values = self.value_projection(values).reshape((B, S, H, -1))
+ out, attn = self.inner_attention(queries, keys, values, attn_mask)
+ out = out.reshape((B, L, -1))
+ return self.out_projection(out), attn
+
+
+class ProbAttention(paddle.nn.Layer):
+ def __init__(
+ self,
+ mask_flag=True,
+ factor=5,
+ scale=None,
+ attention_dropout=0.1,
+ output_attention=False,
+ ):
+ super(ProbAttention, self).__init__()
+ self.factor = factor
+ self.scale = scale
+ self.mask_flag = mask_flag
+ self.output_attention = output_attention
+ self.dropout = paddle.nn.Dropout(p=attention_dropout)
+
+ def _prob_QK(self, Q, K, sample_k, n_top):
+ B, H, L_K, E = tuple(K.shape)
+ _, _, L_Q, _ = tuple(Q.shape)
+ K_expand = K.unsqueeze(axis=-3).expand(shape=[B, H, L_Q, L_K, E])
+ index_sample = paddle.randint(low=0, high=L_K, shape=[L_Q, sample_k])
+ # index_sample = torch.randint(L_K, (L_Q, sample_k))
+ K_sample = K_expand[
+ :, :, paddle.arange(end=L_Q).unsqueeze(axis=1), index_sample, :
+ ]
+
+ x = K_sample
+ perm_5 = list(range(x.ndim))
+ perm_5[-2] = -1
+ perm_5[-1] = -2
+ Q_K_sample = paddle.matmul(
+ x=Q.unsqueeze(axis=-2), y=x.transpose(perm=perm_5)
+ ).squeeze()
+ M = Q_K_sample.max(-1)[0] - paddle.divide(
+ x=Q_K_sample.sum(axis=-1), y=paddle.to_tensor(L_K, dtype="float32")
+ )
+ M_top = M.topk(k=n_top, sorted=False)[1]
+ Q_reduce = Q[
+ paddle.arange(end=B)[:, None, None],
+ paddle.arange(end=H)[None, :, None],
+ M_top,
+ :,
+ ]
+ x = K
+ perm_6 = list(range(x.ndim))
+ perm_6[-2] = -1
+ perm_6[-1] = -2
+ Q_K = paddle.matmul(x=Q_reduce, y=x.transpose(perm=perm_6))
+ return Q_K, M_top
+
+ def _get_initial_context(self, V, L_Q):
+ B, H, L_V, D = tuple(V.shape)
+ if not self.mask_flag:
+ V_sum = V.mean(axis=-2)
+ contex = (
+ V_sum.unsqueeze(axis=-2)
+ .expand(shape=[B, H, L_Q, tuple(V_sum.shape)[-1]])
+ .clone()
+ )
+ else:
+ assert L_Q == L_V
+ contex = V.cumsum(axis=-2)
+ return contex
+
+ def _update_context(self, context_in, V, scores, index, L_Q, attn_mask):
+ B, H, L_V, D = tuple(V.shape)
+ if self.mask_flag:
+ # attn_mask = ProbMask(B, H, L_Q, index, scores, device=V.place)
+ scores.masked_fill_(mask=attn_mask.mask, value=-np.inf)
+ attn = paddle.nn.functional.softmax(x=scores, axis=-1)
+ context_in[
+ paddle.arange(end=B)[:, None, None],
+ paddle.arange(end=H)[None, :, None],
+ index,
+ :,
+ ] = paddle.matmul(x=attn, y=V).astype(dtype=context_in.dtype)
+ if self.output_attention:
+ attns = (
+ (paddle.ones(shape=[B, H, L_Q, L_V]) / L_V)
+ .astype(dtype=attn.dtype)
+ .to(attn.place)
+ )
+ attns[
+ paddle.arange(end=B)[:, None, None],
+ paddle.arange(end=H)[None, :, None],
+ index,
+ :,
+ ] = attn
+ return context_in, attns
+ else:
+ return context_in, None
+
+ def forward(self, queries, keys, values, attn_mask):
+ B, L_Q, H, D = tuple(queries.shape)
+ _, L_K, _, _ = tuple(keys.shape)
+ x = queries
+ perm_7 = list(range(x.ndim))
+ perm_7[2] = 1
+ perm_7[1] = 2
+ queries = x.transpose(perm=perm_7)
+ x = keys
+ perm_8 = list(range(x.ndim))
+ perm_8[2] = 1
+ perm_8[1] = 2
+ keys = x.transpose(perm=perm_8)
+ x = values
+ perm_9 = list(range(x.ndim))
+ perm_9[2] = 1
+ perm_9[1] = 2
+ values = x.transpose(perm=perm_9)
+ U_part = self.factor * np.ceil(np.log(L_K)).astype("int").item()
+ u = self.factor * np.ceil(np.log(L_Q)).astype("int").item()
+ U_part = U_part if U_part < L_K else L_K
+ u = u if u < L_Q else L_Q
+ scores_top, index = self._prob_QK(queries, keys, sample_k=U_part, n_top=u)
+ scale = self.scale or 1.0 / sqrt(D)
+ if scale is not None:
+ scores_top = scores_top * scale
+ context = self._get_initial_context(values, L_Q)
+ context, attn = self._update_context(
+ context, values, scores_top, index, L_Q, attn_mask
+ )
+ return context, attn
+
+
+def FFT_for_Period(x, k=2):
+ xf = paddle.fft.rfft(x=x, axis=1)
+ frequency_list = paddle.abs(xf).mean(axis=0).mean(axis=-1)
+ frequency_list[0] = 0
+ _, top_list = paddle.topk(k=k, x=frequency_list)
+ top_list = top_list.detach().cpu().numpy()
+ period = tuple(x.shape)[1] // top_list
+ return period, paddle.index_select(
+ paddle.abs(xf).mean(axis=-1), paddle.to_tensor(top_list), axis=1
+ )
+
+
+class TimesBlock(paddle.nn.Layer):
+ """Times Block for Spatio-Temporal Attention Fusion Network.
+
+ Args:
+ seq_len (int): Sequence length.
+ pred_len (int): Prediction length.
+ top_k (int): Number of top frequencies to consider.
+ num_kernels (int): Number of kernels in Inception block.
+ d_model (int): Dimension of the model.
+ d_ff (int): Dimension of feedforward network.
+
+ Examples:
+ >>> from ppsci.arch.stafnet import TimesBlock
+ >>> block = TimesBlock(seq_len=72, pred_len=48, top_k=2, num_kernels=32, d_model=512, d_ff=2048)
+ >>> x = paddle.rand([32, 72, 512]) # [batch_size, seq_len, d_model]
+ >>> output = block(x)
+ >>> print(output.shape)
+ [32, 120, 512]
+ """
+
+ def __init__(
+ self,
+ seq_len: int,
+ pred_len: int,
+ top_k: int,
+ num_kernels: int,
+ d_model: int,
+ d_ff: int,
+ ):
+ super(TimesBlock, self).__init__()
+ self.seq_len = seq_len
+ self.pred_len = pred_len
+ self.k = top_k
+ self.conv = paddle.nn.Sequential(
+ Inception_Block_V1(d_model, d_ff, num_kernels=num_kernels),
+ paddle.nn.GELU(),
+ Inception_Block_V1(d_ff, d_model, num_kernels=num_kernels),
+ )
+
+ def forward(self, x):
+ B, T, N = tuple(x.shape)
+ period_list, period_weight = FFT_for_Period(x, self.k)
+ res = []
+ for i in range(self.k):
+ period = period_list[i]
+ if (self.seq_len + self.pred_len) % period != 0:
+ length = ((self.seq_len + self.pred_len) // period + 1) * period
+ padding = paddle.zeros(
+ shape=[
+ tuple(x.shape)[0],
+ length - (self.seq_len + self.pred_len),
+ tuple(x.shape)[2],
+ ]
+ )
+ out = paddle.concat(x=[x, padding], axis=1)
+ else:
+ length = self.seq_len + self.pred_len
+ out = x
+ out = out.reshape((B, length // period, period, N)).transpose(
+ perm=[0, 3, 1, 2]
+ )
+ out = self.conv(out)
+ out = out.transpose(perm=[0, 2, 3, 1]).reshape((B, -1, N))
+ res.append(out[:, : self.seq_len + self.pred_len, :])
+ res = paddle.stack(x=res, axis=-1)
+ period_weight_raw = period_weight
+ period_weight = paddle.nn.functional.softmax(x=period_weight, axis=1)
+ period_weight = paddle.tile(
+ period_weight.unsqueeze(axis=1).unsqueeze(axis=1), (1, T, N, 1)
+ )
+ res = paddle.sum(x=res * period_weight, axis=-1)
+ res = res + x
+ return res, period_list, period_weight_raw
+
+
+def compared_version(ver1, ver2):
+ """
+ :param ver1
+ :param ver2
+ :return: ver1< = >ver2 False/True
+ """
+ list1 = str(ver1).split(".")
+ list2 = str(ver2).split(".")
+ for i in range(len(list1)) if len(list1) < len(list2) else range(len(list2)):
+ if int(list1[i]) == int(list2[i]):
+ pass
+ elif int(list1[i]) < int(list2[i]):
+ return -1
+ else:
+ return 1
+ if len(list1) == len(list2):
+ return True
+ elif len(list1) < len(list2):
+ return False
+ else:
+ return True
+
+
+class PositionalEmbedding(paddle.nn.Layer):
+ def __init__(self, d_model, max_len=5000):
+ super(PositionalEmbedding, self).__init__()
+ pe = paddle.zeros(shape=[max_len, d_model]).astype(dtype="float32")
+ pe.stop_gradient = True
+ position = (
+ paddle.arange(start=0, end=max_len)
+ .astype(dtype="float32")
+ .unsqueeze(axis=1)
+ )
+ div_term = (
+ paddle.arange(start=0, end=d_model, step=2).astype(dtype="float32")
+ * -(math.log(10000.0) / d_model)
+ ).exp()
+ pe[:, 0::2] = paddle.sin(x=position * div_term)
+ pe[:, 1::2] = paddle.cos(x=position * div_term)
+ pe = pe.unsqueeze(axis=0)
+ self.register_buffer(name="pe", tensor=pe)
+
+ def forward(self, x):
+ return self.pe[:, : x.shape[1]]
+
+
+class TokenEmbedding(paddle.nn.Layer):
+ def __init__(self, c_in, d_model):
+ super(TokenEmbedding, self).__init__()
+ padding = 1 if compared_version(paddle.__version__, "1.5.0") else 2
+ self.tokenConv = paddle.nn.Conv1D(
+ in_channels=c_in,
+ out_channels=d_model,
+ kernel_size=3,
+ padding=padding,
+ padding_mode="circular",
+ bias_attr=False,
+ )
+ for m in self.sublayers():
+ if isinstance(m, paddle.nn.Conv1D):
+ init_KaimingNormal = paddle.nn.initializer.KaimingNormal(
+ nonlinearity="leaky_relu"
+ )
+ init_KaimingNormal(m.weight)
+
+ def forward(self, x):
+ x = self.tokenConv(x.transpose(perm=[0, 2, 1]))
+ perm_13 = list(range(x.ndim))
+ perm_13[1] = 2
+ perm_13[2] = 1
+ x = x.transpose(perm=perm_13)
+ return x
+
+
+class FixedEmbedding(paddle.nn.Layer):
+ def __init__(self, c_in, d_model):
+ super(FixedEmbedding, self).__init__()
+ w = paddle.zeros(shape=[c_in, d_model]).astype(dtype="float32")
+ w.stop_gradient = True
+ position = (
+ paddle.arange(start=0, end=c_in).astype(dtype="float32").unsqueeze(axis=1)
+ )
+ div_term = (
+ paddle.arange(start=0, end=d_model, step=2).astype(dtype="float32")
+ * -(math.log(10000.0) / d_model)
+ ).exp()
+ w[:, 0::2] = paddle.sin(x=position * div_term)
+ w[:, 1::2] = paddle.cos(x=position * div_term)
+ self.emb = paddle.nn.Embedding(num_embeddings=c_in, embedding_dim=d_model)
+ out_3 = paddle.create_parameter(
+ shape=w.shape,
+ dtype=w.numpy().dtype,
+ default_initializer=paddle.nn.initializer.Assign(w),
+ )
+ out_3.stop_gradient = not False
+ self.emb.weight = out_3
+
+ def forward(self, x):
+ return self.emb(x).detach()
+
+
+class TemporalEmbedding(paddle.nn.Layer):
+ def __init__(self, d_model, embed_type="fixed", freq="h"):
+ super(TemporalEmbedding, self).__init__()
+ minute_size = 4
+ hour_size = 24
+ weeknum_size = 53
+ weekday_size = 7
+ day_size = 32
+ month_size = 13
+ Embed = FixedEmbedding if embed_type == "fixed" else paddle.nn.Embedding
+ if freq == "t":
+ self.minute_embed = Embed(minute_size, d_model)
+ self.hour_embed = Embed(hour_size, d_model)
+ self.weekday_embed = Embed(weekday_size, d_model)
+ self.weeknum_embed = Embed(weeknum_size, d_model)
+ self.day_embed = Embed(day_size, d_model)
+ self.month_embed = Embed(month_size, d_model)
+ self.Temporal_feature = ["month", "day", "week", "weekday", "hour"]
+
+ def forward(self, x):
+ x = x.astype(dtype="int64")
+ for idx, freq in enumerate(self.Temporal_feature):
+ if freq == "year":
+ pass
+ elif freq == "month":
+ month_x = self.month_embed(x[:, :, idx])
+ elif freq == "day":
+ day_x = self.day_embed(x[:, :, idx])
+ elif freq == "week":
+ weeknum_x = self.weeknum_embed(x[:, :, idx])
+ elif freq == "weekday":
+ weekday_x = self.weekday_embed(x[:, :, idx])
+ elif freq == "hour":
+ hour_x = self.hour_embed(x[:, :, idx])
+ return hour_x + weekday_x + weeknum_x + day_x + month_x
+
+
+class TimeFeatureEmbedding(paddle.nn.Layer):
+ def __init__(self, d_model, embed_type="timeF", freq="h"):
+ super(TimeFeatureEmbedding, self).__init__()
+ freq_map = {"h": 4, "t": 5, "s": 6, "m": 1, "a": 1, "w": 2, "d": 3, "b": 3}
+ d_inp = freq_map[freq]
+ self.embed = paddle.nn.Linear(
+ in_features=d_inp, out_features=d_model, bias_attr=False
+ )
+
+ def forward(self, x):
+ return self.embed(x)
+
+
+class DataEmbedding(paddle.nn.Layer):
+ def __init__(self, c_in, d_model, embed_type="fixed", freq="h", dropout=0.1):
+ super(DataEmbedding, self).__init__()
+ self.value_embedding = TokenEmbedding(c_in=c_in, d_model=d_model)
+ self.position_embedding = PositionalEmbedding(d_model=d_model)
+ self.temporal_embedding = (
+ TemporalEmbedding(d_model=d_model, embed_type=embed_type, freq=freq)
+ if embed_type != "timeF"
+ else TimeFeatureEmbedding(d_model=d_model, embed_type=embed_type, freq=freq)
+ )
+ self.dropout = paddle.nn.Dropout(p=dropout)
+
+ def forward(self, x, x_mark):
+ x = self.value_embedding(
+ x
+ ) # + self.temporal_embedding(x_mark) + self.position_embedding(x)
+ return self.dropout(x)
+
+
+class DataEmbedding_wo_pos(paddle.nn.Layer):
+ def __init__(self, c_in, d_model, embed_type="fixed", freq="h", dropout=0.1):
+ super(DataEmbedding_wo_pos, self).__init__()
+ self.value_embedding = TokenEmbedding(c_in=c_in, d_model=d_model)
+ self.position_embedding = PositionalEmbedding(d_model=d_model)
+ self.temporal_embedding = (
+ TemporalEmbedding(d_model=d_model, embed_type=embed_type, freq=freq)
+ if embed_type != "timeF"
+ else TimeFeatureEmbedding(d_model=d_model, embed_type=embed_type, freq=freq)
+ )
+ self.dropout = paddle.nn.Dropout(p=dropout)
+
+ def forward(self, x, x_mark):
+ x = self.value_embedding(x) + self.temporal_embedding(x_mark)
+ return self.dropout(x)
+
+
+class GAT_Encoder(paddle.nn.Layer):
+ def __init__(self, input_dim, hid_dim, edge_dim, gnn_embed_dim, dropout):
+ super(GAT_Encoder, self).__init__()
+ self.input_dim = input_dim
+ self.hid_dim = hid_dim
+ self.relu = paddle.nn.ReLU()
+ self.dropout = paddle.nn.Dropout(p=dropout)
+ self.conv1 = GATv2Conv(
+ input_dim,
+ hid_dim,
+ )
+ self.conv2 = GATv2Conv(
+ hid_dim,
+ hid_dim * 2,
+ )
+ self.conv3 = GATv2Conv(
+ hid_dim * 2,
+ gnn_embed_dim,
+ )
+
+ def forward(
+ self,
+ graph,
+ feature,
+ ):
+ x = self.conv1(graph, feature)
+ x = self.relu(x)
+ # x = x.relu()
+ x = self.conv2(graph, x)
+ x = self.relu(x)
+ x = self.conv3(graph, x)
+ x = self.dropout(x)
+
+ return x
+
+
+class STAFNet(base.Arch):
+ """Spatio-Temporal Attention Fusion Network (STAFNet).
+
+ STAFNet is a neural network architecture for spatio-temporal data prediction, combining attention mechanisms and convolutional neural networks to capture spatio-temporal dependencies.
+
+ Args:
+ input_keys (Tuple[str, ...]): Keys of input variables.
+ output_keys (Tuple[str, ...]): Keys of output variables.
+ seq_len (int): Sequence length.
+ pred_len (int): Prediction length.
+ aq_gat_node_num (int): Number of nodes in the air quality GAT.
+ aq_gat_node_features (int): Number of features for each node in the air quality GAT.
+ mete_gat_node_num (int): Number of nodes in the meteorological GAT.
+ mete_gat_node_features (int): Number of features for each node in the meteorological GAT.
+ gat_hidden_dim (int): Hidden dimension of the GAT.
+ gat_edge_dim (int): Edge dimension of the GAT.
+ d_model (int): Dimension of the model.
+ n_heads (int): Number of attention heads.
+ e_layers (int): Number of encoder layers.
+ enc_in (int): Encoder input dimension.
+ dec_in (int): Decoder input dimension.
+ freq (str): Frequency for positional encoding.
+ embed (str): Embedding type.
+ d_ff (int): Dimension of feedforward network.
+ top_k (int): Number of top frequencies to consider.
+ num_kernels (int): Number of kernels in Inception block.
+ dropout (float): Dropout rate.
+ output_attention (bool): Whether to output attention.
+ factor (int): Factor for attention mechanism.
+ c_out (int): Output channels.
+
+ Examples:
+ >>> from ppsci.arch import STAFNet
+ >>> model = STAFNet(
+ ... input_keys=('x', 'y', 'z'),
+ ... output_keys=('u', 'v'),
+ ... seq_len=72,
+ ... pred_len=48,
+ ... aq_gat_node_num=10,
+ ... aq_gat_node_features=16,
+ ... mete_gat_node_num=10,
+ ... mete_gat_node_features=16,
+ ... gat_hidden_dim=32,
+ ... gat_edge_dim=8,
+ ... d_model=512,
+ ... n_heads=8,
+ ... e_layers=3,
+ ... enc_in=7,
+ ... dec_in=7,
+ ... freq='h',
+ ... embed='fixed',
+ ... d_ff=2048,
+ ... top_k=5,
+ ... num_kernels=32,
+ ... dropout=0.1,
+ ... output_attention=False,
+ ... factor=5,
+ ... c_out=7,
+ ... )
+ >>> input_dict = {"x": paddle.rand([32, 72, 7]),
+ ... "y": paddle.rand([32, 72, 7]),
+ ... "z": paddle.rand([32, 72, 7])}
+ >>> output_dict = model(input_dict)
+ >>> print(output_dict["u"].shape)
+ [32, 48, 7]
+ >>> print(output_dict["v"].shape)
+ [32, 48, 7]
+ """
+
+ def __init__(
+ self,
+ input_keys: Tuple[str, ...],
+ output_keys: Tuple[str, ...],
+ seq_len: int,
+ pred_len: int,
+ aq_gat_node_num: int,
+ aq_gat_node_features: int,
+ mete_gat_node_num: int,
+ mete_gat_node_features: int,
+ gat_hidden_dim: int,
+ gat_edge_dim: int,
+ d_model: int,
+ n_heads: int,
+ e_layers: int,
+ enc_in: int,
+ dec_in: int,
+ freq: str,
+ embed: str,
+ d_ff: int,
+ top_k: int,
+ num_kernels: int,
+ dropout: float,
+ output_attention: bool,
+ factor: int,
+ c_out: int,
+ ):
+ super(STAFNet, self).__init__()
+ self.input_keys = input_keys
+ self.output_keys = output_keys
+ self.device = str("cuda").replace("cuda", "gpu")
+ self.output_attention = output_attention
+ self.seq_len = seq_len
+ self.pred_len = pred_len
+ self.dec_in = dec_in
+ self.gat_embed_dim = enc_in
+ self.enc_embedding = DataEmbedding(enc_in, d_model, embed, freq, dropout)
+ self.aq_gat_node_num = aq_gat_node_num
+ self.aq_gat_node_features = aq_gat_node_features
+ self.aq_GAT = GAT_Encoder(
+ aq_gat_node_features,
+ gat_hidden_dim,
+ gat_edge_dim,
+ self.gat_embed_dim,
+ dropout,
+ ).to(self.device)
+ self.mete_gat_node_num = mete_gat_node_num
+ self.mete_gat_node_features = mete_gat_node_features
+ self.mete_GAT = GAT_Encoder(
+ mete_gat_node_features,
+ gat_hidden_dim,
+ gat_edge_dim,
+ self.gat_embed_dim,
+ dropout,
+ ).to(self.device)
+ self.pos_fc = paddle.nn.Linear(
+ in_features=2, out_features=self.gat_embed_dim, bias_attr=True
+ )
+ self.fusion_Attention = AttentionLayer(
+ ProbAttention(
+ False,
+ factor,
+ attention_dropout=dropout,
+ output_attention=self.output_attention,
+ ),
+ self.gat_embed_dim,
+ n_heads,
+ )
+ self.model = paddle.nn.LayerList(
+ sublayers=[
+ TimesBlock(seq_len, pred_len, top_k, num_kernels, d_model, d_ff)
+ for _ in range(e_layers)
+ ]
+ )
+ self.layer = e_layers
+ self.layer_norm = paddle.nn.LayerNorm(normalized_shape=d_model)
+ self.predict_linear = paddle.nn.Linear(
+ in_features=self.seq_len, out_features=self.pred_len + self.seq_len
+ )
+ self.projection = paddle.nn.Linear(
+ in_features=d_model, out_features=c_out, bias_attr=True
+ )
+ self.output_attention = output_attention
+
+ def aq_gat(self, G):
+ g_batch = G.num_graph
+ batch_size = int(g_batch / self.seq_len)
+ gat_output = self.aq_GAT(
+ G, G.node_feat["feature"][:, -self.aq_gat_node_features :]
+ )
+ gat_output = gat_output.reshape(
+ (batch_size, self.seq_len, self.aq_gat_node_num, self.gat_embed_dim)
+ )
+ gat_output = paddle.flatten(x=gat_output, start_axis=0, stop_axis=1)
+ return gat_output
+
+ def mete_gat(self, G):
+ g_batch = G.num_graph
+ batch_size = int(g_batch / self.seq_len)
+ gat_output = self.mete_GAT(
+ G, G.node_feat["feature"][:, -self.mete_gat_node_features :]
+ )
+ gat_output = gat_output.reshape(
+ (batch_size, self.seq_len, self.mete_gat_node_num, self.gat_embed_dim)
+ )
+ gat_output = paddle.flatten(x=gat_output, start_axis=0, stop_axis=1)
+ return gat_output
+
+ def norm_pos(self, A, B):
+ A_mean = paddle.mean(A, axis=0)
+ A_std = paddle.std(A, axis=0)
+
+ A_norm = (A - A_mean) / A_std
+ B_norm = (B - A_mean) / A_std
+ return A_norm, B_norm
+
+ def forward(self, Data, mask=None):
+ train_data = Data["aq_train_data"]
+ batch_size = train_data.shape[0]
+ x = train_data
+ perm_1 = list(range(x.ndim))
+ perm_1[1] = 2
+ perm_1[2] = 1
+ train_data = paddle.transpose(x=x, perm=perm_1)
+ train_data = paddle.flatten(x=train_data, start_axis=0, stop_axis=1)
+ x_enc = train_data[:, : self.seq_len, -self.dec_in :]
+ x_mark_enc = train_data[:, : self.seq_len, 1:6]
+
+ means = x_enc.mean(axis=1, keepdim=True).detach()
+ x_enc = x_enc - means
+ stdev = paddle.sqrt(
+ x=paddle.var(x=x_enc, axis=1, keepdim=True, unbiased=False) + 1e-05
+ )
+ x_enc /= stdev
+ # enc_out = self.enc_embedding(aq_gat_output, x_mark_enc)
+ enc_out = self.enc_embedding(x_enc, x_mark_enc)
+ enc_out = self.predict_linear(enc_out.transpose(perm=[0, 2, 1])).transpose(
+ perm=[0, 2, 1]
+ )
+ for i in range(self.layer):
+ enc_out, period_list, period_weight = self.model[i](enc_out)
+ enc_out = self.layer_norm(enc_out)
+ dec_out = self.projection(enc_out)
+ dec_out = dec_out * paddle.tile(
+ stdev[:, 0, :].unsqueeze(axis=1), (1, self.pred_len + self.seq_len, 1)
+ )
+ dec_out = dec_out + paddle.tile(
+ means[:, 0, :].unsqueeze(axis=1), (1, self.pred_len + self.seq_len, 1)
+ )
+
+ return {
+ self.output_keys[0]: dec_out[:, -self.pred_len :, -7:].reshape(
+ (batch_size, self.pred_len, -1, 7)
+ )
+ }
diff --git a/ppsci/data/dataset/__init__.py b/ppsci/data/dataset/__init__.py
index 9ece354700..bd10c49680 100644
--- a/ppsci/data/dataset/__init__.py
+++ b/ppsci/data/dataset/__init__.py
@@ -46,6 +46,7 @@
from ppsci.data.dataset.radar_dataset import RadarDataset
from ppsci.data.dataset.sevir_dataset import SEVIRDataset
from ppsci.data.dataset.spherical_swe_dataset import SphericalSWEDataset
+from ppsci.data.dataset.stafnet_dataset import STAFNetDataset
from ppsci.data.dataset.trphysx_dataset import CylinderDataset
from ppsci.data.dataset.trphysx_dataset import LorenzDataset
from ppsci.data.dataset.trphysx_dataset import RosslerDataset
@@ -93,6 +94,7 @@
"DrivAerNetDataset",
"DrivAerNetPlusPlusDataset",
"IFMMoeDataset",
+ "STAFNetDataset",
]
diff --git a/ppsci/data/dataset/stafnet_dataset.py b/ppsci/data/dataset/stafnet_dataset.py
new file mode 100644
index 0000000000..e61ebbfa89
--- /dev/null
+++ b/ppsci/data/dataset/stafnet_dataset.py
@@ -0,0 +1,180 @@
+from typing import Optional
+from typing import Tuple
+
+import numpy as np
+import paddle
+import pandas
+
+try:
+ import pgl
+except ModuleNotFoundError:
+ pass
+from paddle import io
+from paddle.io import DataLoader
+from scipy.spatial.distance import cdist
+
+
+def gat_lstmcollate_fn(data):
+ aq_train_data = []
+ mete_train_data = []
+ aq_g_list = []
+ mete_g_list = []
+ label = []
+ for unit in data:
+ aq_train_data.append(unit[0]["aq_train_data"])
+ mete_train_data.append(unit[0]["mete_train_data"])
+ aq_g_list = aq_g_list + unit[0]["aq_g_list"]
+ mete_g_list = mete_g_list + unit[0]["mete_g_list"]
+ label.append(unit[1])
+ label = paddle.stack(x=label)
+ x = label
+ perm_1 = list(range(x.ndim))
+ perm_1[1] = 2
+ perm_1[2] = 1
+ label = paddle.transpose(x=x, perm=perm_1)
+ label = paddle.flatten(x=label, start_axis=0, stop_axis=1)
+ return {
+ "aq_train_data": paddle.stack(x=aq_train_data),
+ "mete_train_data": paddle.stack(x=mete_train_data),
+ "aq_G": pgl.graph.Graph.batch(aq_g_list),
+ "mete_G": pgl.graph.Graph.batch(mete_g_list),
+ }, label
+
+
+class pygmmdataLoader(DataLoader):
+ """
+ MNIST data loading demo using BaseDataLoader
+ """
+
+ def __init__(
+ self,
+ args,
+ data_dir,
+ batch_size,
+ shuffle=True,
+ num_workers=1,
+ training=True,
+ T=24,
+ t=12,
+ collate_fn=gat_lstmcollate_fn,
+ ):
+ self.T = T
+ self.t = t
+ self.dataset = STAFNetDataset(args=args, file_path=data_dir)
+
+ super().__init__(
+ self.dataset,
+ batch_size=batch_size,
+ shuffle=shuffle,
+ num_workers=num_workers,
+ collate_fn=collate_fn,
+ )
+
+
+class STAFNetDataset(io.Dataset):
+ """Dataset class for STAFNet data.
+
+ Args:
+ file_path (str): Path to the dataset file.
+ input_keys (Optional[Tuple[str, ...]]): Tuple of input keys. Defaults to None.
+ label_keys (Optional[Tuple[str, ...]]): Tuple of label keys. Defaults to None.
+ seq_len (int): Sequence length. Defaults to 72.
+ pred_len (int): Prediction length. Defaults to 48.
+ use_edge_attr (bool): Whether to use edge attributes. Defaults to True.
+
+ Examples:
+ >>> from ppsci.data.dataset import STAFNetDataset
+
+ >>> dataset = STAFNetDataset(file_path='example.pkl') # doctest: +SKIP
+
+ >>> # get the length of the dataset
+ >>> dataset_size = len(dataset) # doctest: +SKIP
+ >>> # get the first sample of the data
+ >>> first_sample = dataset[0] # doctest: +SKIP
+ >>> print("First sample:", first_sample) # doctest: +SKIP
+ """
+
+ def __init__(
+ self,
+ file_path: str,
+ input_keys: Optional[Tuple[str, ...]] = None,
+ label_keys: Optional[Tuple[str, ...]] = None,
+ seq_len: int = 72,
+ pred_len: int = 48,
+ use_edge_attr: bool = True,
+ ):
+
+ self.file_path = file_path
+ self.input_keys = input_keys
+ self.label_keys = label_keys
+ self.use_edge_attr = use_edge_attr
+
+ self.seq_len = seq_len
+ self.pred_len = pred_len
+
+ super().__init__()
+ if file_path.endswith(".pkl"):
+ with open(file_path, "rb") as f:
+ self.data = pandas.read_pickle(f)
+ self.metedata = self.data["metedata"]
+ self.AQdata = self.data["AQdata"]
+ self.AQStation_imformation = self.data["AQStation_imformation"]
+ self.meteStation_imformation = self.data["meteStation_imformation"]
+ mete_coords = np.array(
+ self.meteStation_imformation.loc[:, ["经度", "纬度"]]
+ ).astype("float64")
+ AQ_coords = np.array(self.AQStation_imformation.iloc[:, -2:]).astype("float64")
+ self.aq_edge_index, self.aq_edge_attr, self.aq_node_coords = self.get_edge_attr(
+ np.array(self.AQStation_imformation.iloc[:, -2:]).astype("float64")
+ )
+ (
+ self.mete_edge_index,
+ self.mete_edge_attr,
+ self.mete_node_coords,
+ ) = self.get_edge_attr(
+ np.array(self.meteStation_imformation.loc[:, ["经度", "纬度"]]).astype(
+ "float64"
+ )
+ )
+
+ self.lut = self.find_nearest_point(AQ_coords, mete_coords)
+
+ def __len__(self):
+ return len(self.AQdata) - self.seq_len - self.pred_len
+
+ def __getitem__(self, idx):
+ aq_train_data = paddle.to_tensor(
+ data=self.AQdata[idx : idx + self.seq_len + self.pred_len]
+ ).astype(dtype="float32")
+ mete_train_data = paddle.to_tensor(
+ data=self.metedata[idx : idx + self.seq_len + self.pred_len]
+ ).astype(dtype="float32")
+
+ input_item = {
+ "aq_train_data": aq_train_data,
+ "mete_train_data": mete_train_data,
+ }
+ label_item = {self.label_keys[0]: aq_train_data[-self.pred_len :, :, -7:]}
+
+ return input_item, label_item, {}
+
+ def get_edge_attr(self, node_coords, threshold=0.2):
+ # node_coords = paddle.to_tensor(data=node_coords)
+ dist_matrix = cdist(node_coords, node_coords)
+ edge_index = np.where(dist_matrix < threshold)
+ # edge_index = paddle.to_tensor(data=edge_index, dtype='int64')
+ start_nodes, end_nodes = edge_index
+ edge_lengths = dist_matrix[start_nodes, end_nodes]
+ edge_directions = node_coords[end_nodes] - node_coords[start_nodes]
+ edge_attr = paddle.to_tensor(
+ data=np.concatenate((edge_lengths[:, np.newaxis], edge_directions), axis=1)
+ )
+ node_coords = paddle.to_tensor(data=node_coords)
+ return edge_index, edge_attr, node_coords
+
+ def find_nearest_point(self, A, B):
+ nearest_indices = []
+ for a in A:
+ distances = [np.linalg.norm(a - b) for b in B]
+ nearest_indices.append(np.argmin(distances))
+ return nearest_indices

