-
|
I'm trying to train a model to segment the BraTs dataset, and I'm using this tutorial as a starting point. My understanding is that augmentation transforms (such as RandFlipd) should be applied randomly BUT equally for all channels in the image (in this case MRI sequences) and the labels. It seems like what happens instead is that the transform is applied randomly to each channel (both of image and label), so that they don't necessary match after the transform. Each sequence is flipped/not flipped independently of each other, and the same happens with the labels. This can be reproduced by running the tutorial changing val_ds for train_ds in 'Check data shape and visualize' (random transforms are only applied to the train dataset, so the validation dataset, which the tutorial plots, is fine). Any tips on how to avoid this are much appreciated! |
Beta Was this translation helpful? Give feedback.
Replies: 1 comment 3 replies
-
|
Hi @lidialuq , Thanks for your interest here. Thanks in advance. |
Beta Was this translation helpful? Give feedback.
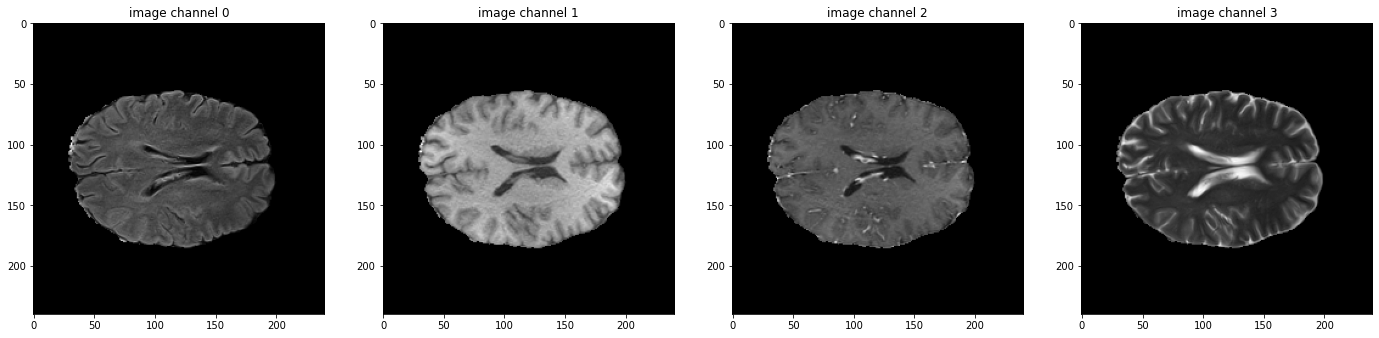
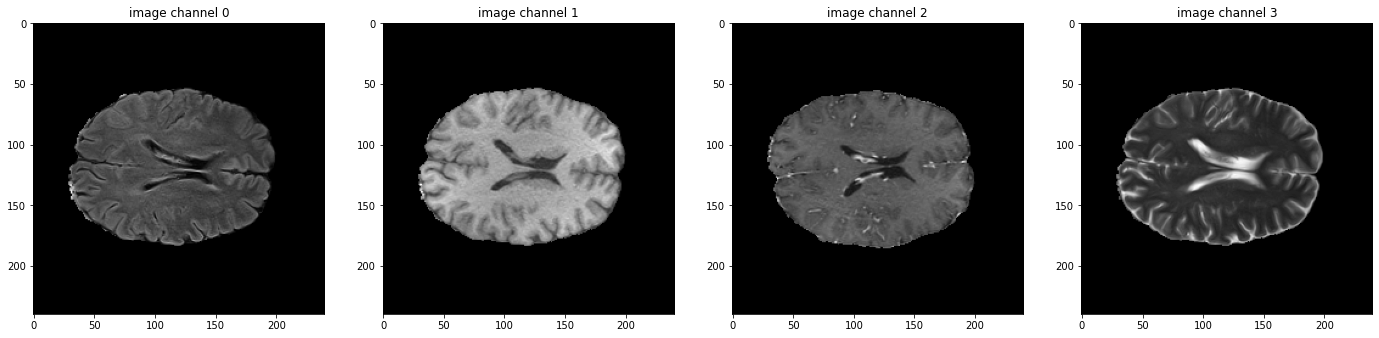
Hi @lidialuq ,
Thanks for your interest here.
I think we applied
map_spatial_axesinRandFlipto flip all the channels together:https://github.com/Project-MONAI/MONAI/blob/dev/monai/transforms/spatial/array.py#L362
Could you please help share some test result images that the random operation on different channel is different?
Thanks in advance.