segmentation has different orientation from original volume #3719
Replies: 3 comments 6 replies
-
|
And what does your visualisation code look like? If you superpose |
Beta Was this translation helpful? Give feedback.
-
|
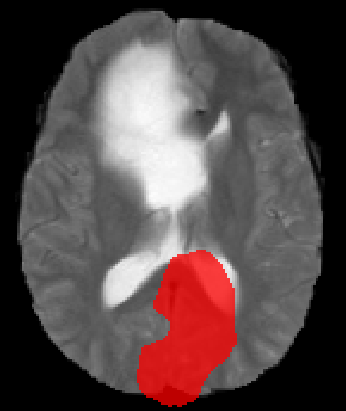
Hi @rijobro! To double check I also plotted them in matplotlib, but it still looks like the orientations are different |
Beta Was this translation helpful? Give feedback.
-
|
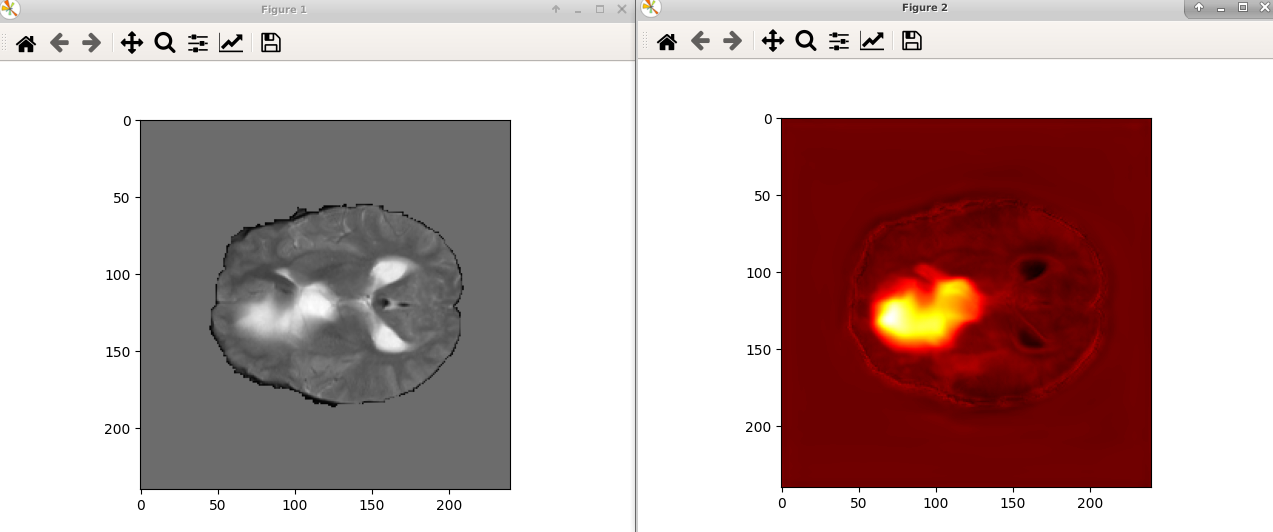
So, the way I created the plot with matplotlib was simply loading the saved mask from disk with: import nibabel as nib
import numpy as np
import matplotlib.pyplot as plt
orig_volume = nib.load("/media/newuser/7EF9B56B27259371/BRATS_dataset/Task01_Brain_Tumour_Reduced/imagesTs/BRATS_487.nii.gz")
orig_volume_np = np.asanyarray(orig_volume.dataobj)
segm_volume = nib.load("/media/newuser/7EF9B56B27259371/BRATS_dataset/out_inference/out_brats_inference_Jan_26_2022/BRATS_487_seg.nii.gz")
segm_volume_np = np.asanyarray(segm_volume.dataobj)
plt.figure(1)
slice = 79
plt.imshow(orig_volume_np[:, :, slice], cmap="gray")
plt.imshow(segm_volume_np[:, :, slice], alpha=0.5)
plt.show()but you are right, if I plot them before thresholding and before saving to disk, they are aligned! I tried with: for test_data in test_loader: # loop over batches
test_inputs = test_data["image"].to(device) # type: torch.Tensor # extract volumes
test_data["pred"] = inference(test_inputs, val_amp, model) # compute inference
slice = 79
test_inputs_np = test_inputs.cpu().detach().numpy()[0, 0, :, :, slice]
test_pred_np = test_data["pred"].cpu().detach().numpy()[0, 0, :, :, slice]
plt.figure(1)
plt.imshow(test_inputs_np, cmap="gray")
plt.figure(2)
plt.imshow(test_pred_np, cmap="hot")So I guess the problem is in the saving process (i.e. when I call |
Beta Was this translation helpful? Give feedback.
Uh oh!
There was an error while loading. Please reload this page.
-
Hi :)
I am working on an adapted version of the BRATS 3D segmentation script. Training works smoothly and the validation dice looks fine. Now, I wanted run inference on the test set and save the segmentation masks to disk.
I’ve been following tutorial1 and tutorial2
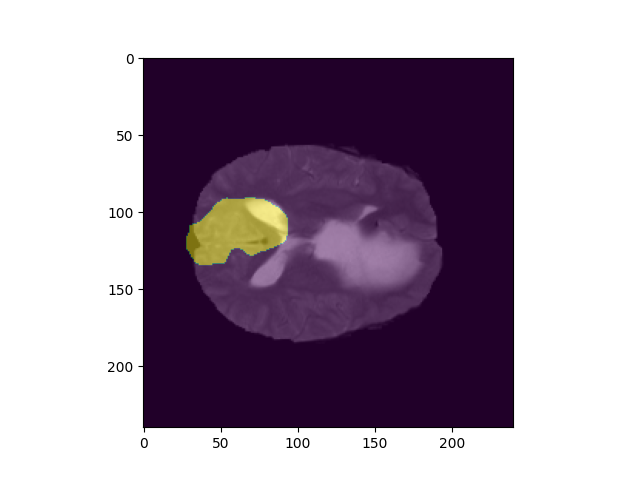
However, it looks like the segmentations are correct but have a different orientation from the original volumes (see image)
What am I doing wrong? Thanks a lot in advance! here is my pseudo code:
Beta Was this translation helpful? Give feedback.
All reactions