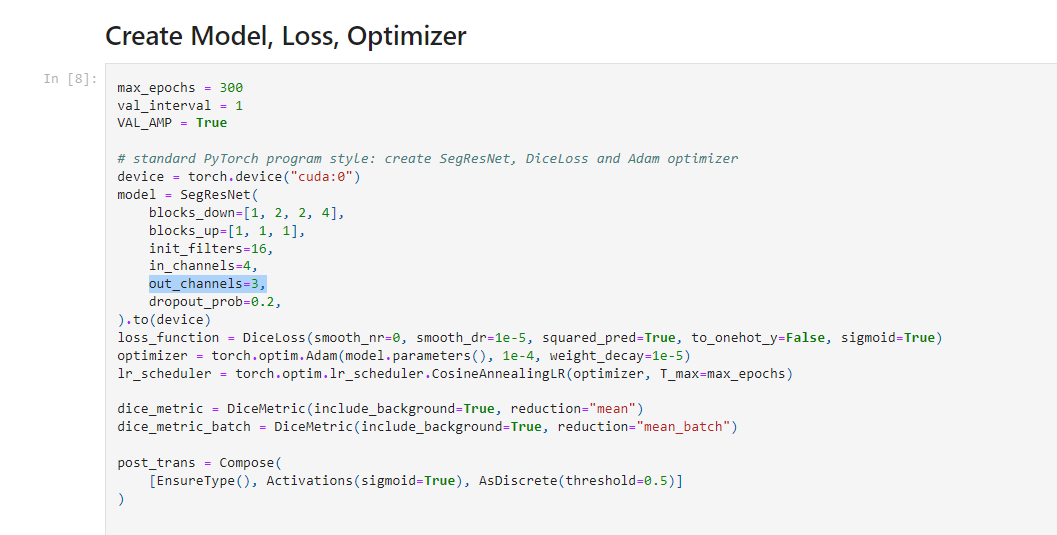
out_channels in tutorial:"Brain tumor 3D segmentation with MONAI" should be 4 #4022
-
|
For multi-label segmentation, it has been said (#415 (comment)) that |
Beta Was this translation helpful? Give feedback.
Replies: 3 comments 1 reply
-
|
Hi @ankitabuntolia , I think we don't have the background channel in the label of brats dataset, so set the output of SegResNet to 3. So we will not predict the background at all. Thanks. |
Beta Was this translation helpful? Give feedback.
-
|
In my task, I have 4 unique values in labels (0, 1, 2, 3) where 0=Background, 1, 2, 3 are eye fluids. The input image and label size are 512x496x49. I followed the same method (as in BRAT tutorial ) to convert labels to multi-channels. thus after transforming [spatial_size (128x128x128) in Rand3DElasticd] the shape of labels are 3 channels (3x512x496x49) and the shape of Images are (1x512x496x49). So Actually in provided labels, I do have 0 as a background label so do you think, I should consider it while using the |
Beta Was this translation helpful? Give feedback.
-
|
I think maybe you can just ignore the background label, same as the brain tumor tutorial, it depends on your needs. Thanks. |
Beta Was this translation helpful? Give feedback.
I think maybe you can just ignore the background label, same as the brain tumor tutorial, it depends on your needs.
@dongyang0122 May help provide more details from the research angle if you have more questions about the channels.
Thanks.