Voxel-wise classification training pipeline #5318
-
|
I want to train simple neural network two layer classifier. Input data is 3-dimensional volumes of feature maps extracted from Brain MRI. I have manually extracted 6 feature maps each size of [128,128,128]. I want to feed each voxel into neural network, so input layer will have 6 neurons (for each map). The labels are the masks that manually segmented in region of brain malformation. I got following question regarding implementation: The issue is I don’t quite understand how to create a training pipeline and dataloader for this type of input. I assume to create PatchDataset with size 1x1x1 https://docs.monai.io/en/stable/data.html?highlight=patchdataset#patchdataset. Are there examples of voxel-wise training example? I apologies, if this question doesn't relate discussion topics. |
Beta Was this translation helpful? Give feedback.
Replies: 1 comment 6 replies
-
|
it sounds like there's nothing spatial about the model you want to learn? i.e. the prediction for a given voxel should be independent of the features in the other voxels near to it? if so then maybe PatchDataset is overkill and all you need to do is reshape to absorb the spatial dimension into the batch dimension
|
Beta Was this translation helpful? Give feedback.
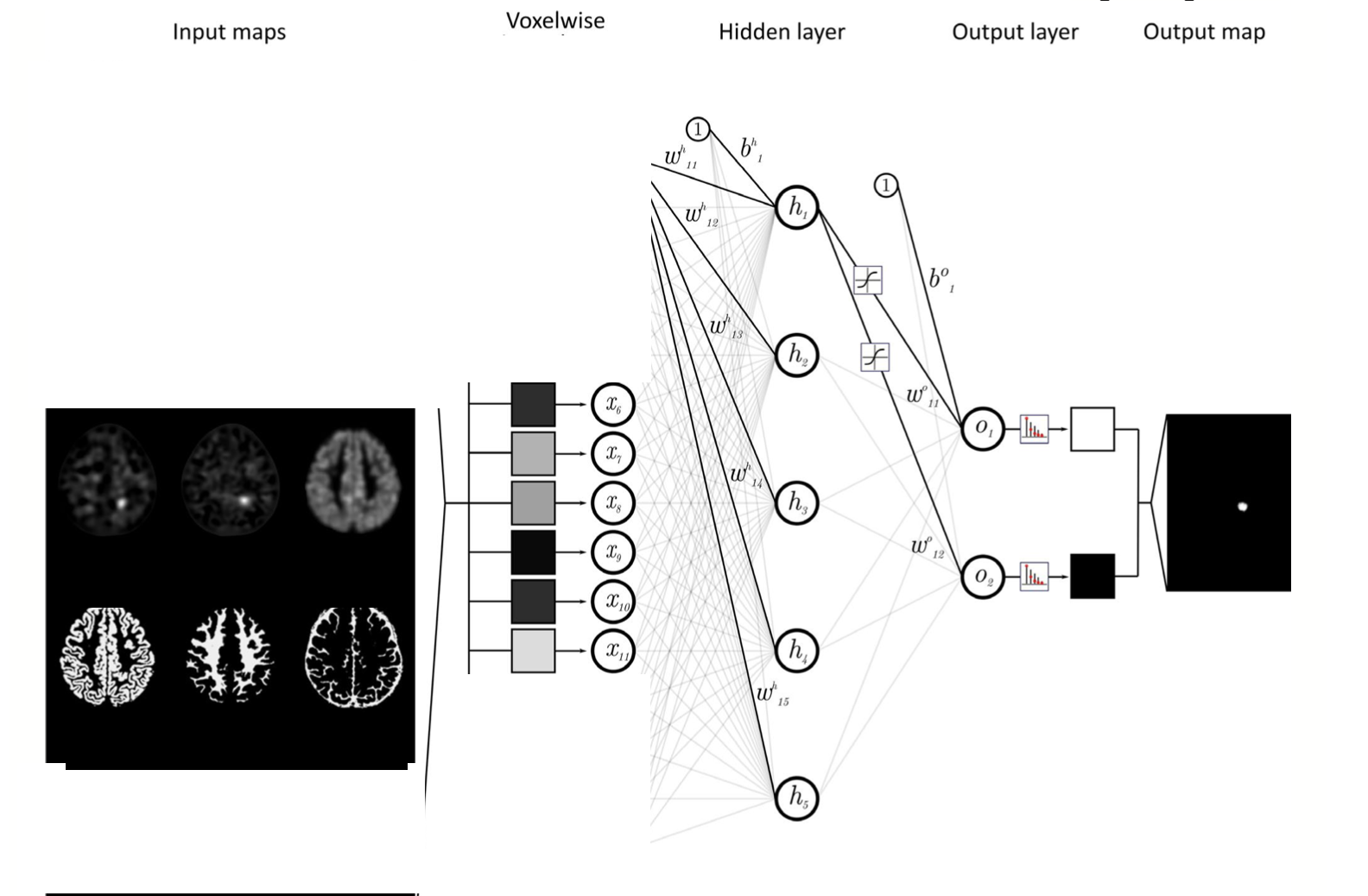
it sounds like there's nothing spatial about the model you want to learn? i.e. the prediction for a given voxel should be independent of the features in the other voxels near to it?
if so then maybe PatchDataset is overkill and all you need to do is reshape to absorb the spatial dimension into the batch dimension
img.permute(0,2,3,4,1)will move the spatial dimensions to be adjacent to batch dimension giving you shapeimg.reshape(-1,6)will then absorb the spatial dimensions into the batch dimension, giving shape