Replies: 1 comment 3 replies
-
|
Hi @aalhayali, 1 in Hope it can help you, thanks! |
Beta Was this translation helpful? Give feedback.
3 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
Hi!
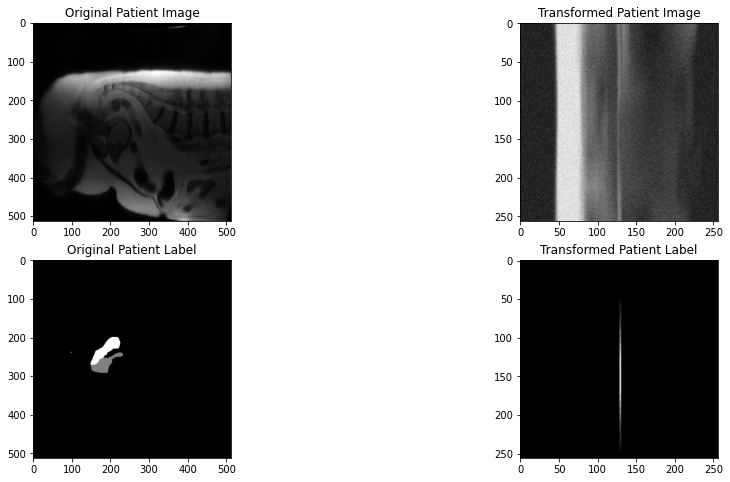
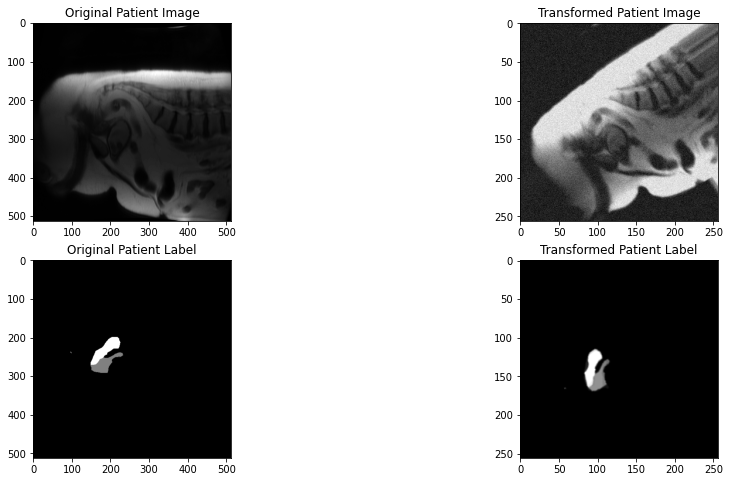
I'm currently training a 3D-UNet model. The image and label size I'm working with are of sagittal MRI
(512,512,7). I'm using the following transforms for both train and test (minus the augmentations of course):The output image size after loading the dataset to a dataloader and using a batch size of 2 is
(2, 1, 7, 512, 512). I I understand that the 1 is a placeholder (though don't fully understand it's purpose, I'd appreciate if someone can clarify it). When passing the dataloaders into the training loop, the image is increasing by 1 in the second dimension (index 1). Here's a snippet of the training code:Image and label shape before passing
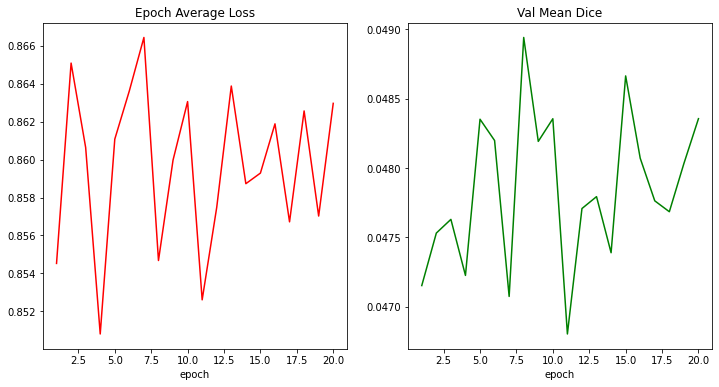
volumetomodelwere (2, 1, 7, 256, 256), however,outputgenerates a prediction with the following shape:(2, 2, 7, 256, 256), while the label's shape remains as it was originally. This of course is generating aValueError: y_pred and y should have same shapes, got (2, 2, 7, 256, 256) and (2, 1, 7, 256, 256).due to the mismatch, precisely at thetrain_lossvariable. May I know why I'm getting this dimensionality increase? I looked around for a while and have yet to see this issue.Thanks!
Beta Was this translation helpful? Give feedback.
All reactions