Replies: 2 comments
-
|
Hi @ahxmeds, since |
Beta Was this translation helpful? Give feedback.
-
|
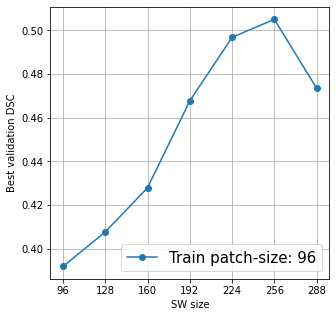
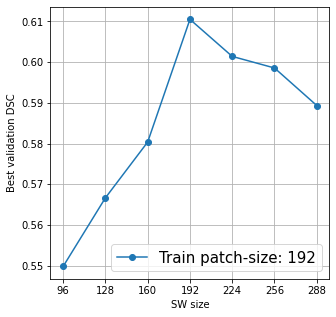
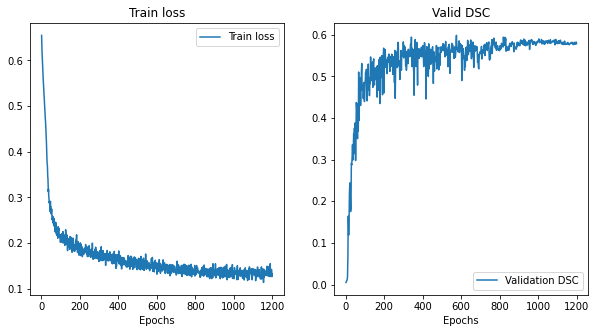
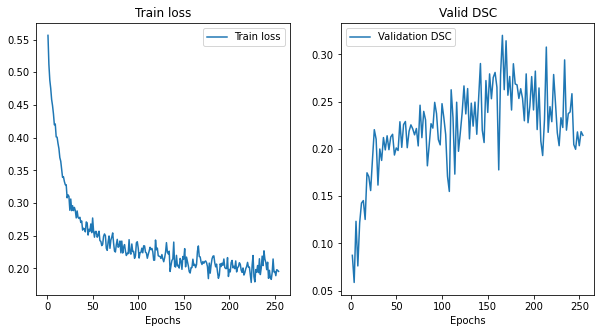
@KumoLiu Thank you. That was helpful, the As you can see in the above plots, for N=96, the optimal M=256, and for N=192, M=192. So, it is evident that the training patch size and the corresponding inference window can strongly influence the model performance and model selection. And, my initial plan was to replicate this ablation experiments in other networks as well, like I also noticed another thing: I used I tried training Training loss and validation DSC for Training loss and validation DSC for Training loss and validation DSC for Again, thanks for your help so far. :) |
Beta Was this translation helpful? Give feedback.

Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
Hi,
I have used the 3D segmentation code (with modification) based on the spleen segmentation code in the MONAI tutorials for my own PET/CT segmentation project. My code seems to work fine when I use
UNetas the network. But if I try using others network (like SwinUNETR or DynUNet), I getRuntimeError: CUDA out of memory. I am only testing this with 2 training and validation PET/CT images for now.My
train_transformandvalid_transformlook like this:And I am doing the
modeldefinition like this:The training for the
UNetmodel (that has been commented out above) seems to work with this code, but using SwinUNETR (or DynUNet) generates this error:Surprisingly, I tried running the whole thing on the CPU and the training runs successfully on the CPU with absolutely no errors (although very slow, taking about 2 mins per epoch, even with 2 training images). Please let me know how to fix this error (as I do not understand it) and help me run my code on the GPU(s).
Beta Was this translation helpful? Give feedback.
All reactions