diff --git a/docs/compute-engine-idle-shutdown.md b/docs/compute-engine-idle-shutdown.md
index 625192f..99bb6ea 100644
--- a/docs/compute-engine-idle-shutdown.md
+++ b/docs/compute-engine-idle-shutdown.md
@@ -2,7 +2,9 @@
This guide walks you through how to automatically configure auto-shutdown capabilities on any GCP virtual machine. Put simply, you only need to add a startup script that shuts down the VM after a specified period of inactivity. This safeguard helps ensure you don't forget to shut off your VM over the weekend, and accidentally burn through a big chunk of your budget.
## 1. Configure auto-shutdown on Vertex AI Notebook Instance
-When starting a new VertexAI notebook, you can enable idle shutdown by simply clicking a box and specifying the amount of time you want your machine idle before it shuts down. See our [VertexAI startup guide](/docs/vertexai.md) to see how to enable this important feature.
+When starting a new VertexAI notebook, you can enable idle shutdown by navigating to **Advanced options** and checking the box for enabling auto-shutdown. You must specify the amount of time you want your machine idle before it shuts down. See our [VertexAI startup guide](/docs/vertexai.md) to see how to enable this important feature.
+
+
For additional details on the other settings for user managed notebooks, see [this VertexAI Quickstart](https://cloud.google.com/vertex-ai/docs/workbench/user-managed/create-user-managed-notebooks-instance-console-quickstart).
diff --git a/docs/idle-shutdown.sh b/docs/idle-shutdown.sh
index 27f9f68..2f3513a 100644
--- a/docs/idle-shutdown.sh
+++ b/docs/idle-shutdown.sh
@@ -4,7 +4,7 @@
# NOTE: requires `bc`, eg, sudo apt-get install bc
# Modified from https://stackoverflow.com/questions/30556920/how-can-i-automatically-kill-idle-gce-instances-based-on-cpu-usage
-sudo apt-get install bc
+sudo apt-get install bc -y
# This is the CPU usage threshold. If activity falls below 10% for the specified time below, the VM shutsdown.
# If you want the shutdown to be more sensitive, you can set this higher, so that a smaller drop in CPU activity will cause shutdown.
diff --git a/docs/protein_setup.md b/docs/protein_setup.md
index a73913f..a5d5785 100644
--- a/docs/protein_setup.md
+++ b/docs/protein_setup.md
@@ -1,7 +1,21 @@
-!curl -L -O https://github.com/conda-forge/miniforge/releases/latest/download/Mambaforge-$(uname)-$(uname -m).sh
+# Setting Up the Environment to Run AlphaFold on Vertex AI
-!bash Mambaforge-$(uname)-$(uname -m).sh -b -p $HOME/mambaforge
+## 1. Install Mamba (via Mambaforge)
+Use the following commands to install Mambaforge:
-!../../mambaforge/bin/mamba install -y -c conda-forge pandas-gbq pandas
+```bash
+# Download the Mambaforge installer
+curl -L -O https://github.com/conda-forge/miniforge/releases/latest/download/Mambaforge-$(uname)-$(uname -m).sh
+# Install Mambaforge to your home directory
+bash Mambaforge-$(uname)-$(uname -m).sh -b -p $HOME/mambaforge
+
+# Use mamba to install required Python packages
+$HOME/mambaforge/bin/mamba install -y -c conda-forge pandas-gbq pandas
+```
+## 2. Install Google Colab Package
+Use the following command to install google-colab:
+
+```bash
pip install google-colab
+```
diff --git a/docs/vertexai.md b/docs/vertexai.md
index 3f38357..a4624b8 100644
--- a/docs/vertexai.md
+++ b/docs/vertexai.md
@@ -1,9 +1,9 @@
# Using VertexAI Notebooks
### 1. Spin up an Instance
-1. Start by clicking the `hamburger menu` (the three horizontal lines in the top left of your console). Go to `Artificial Intelligence > Vertex AI > Workbench`.
+1. Start by clicking the `hamburger menu` (the three horizontal lines in the top left of your console). Go to `Vertex AI > Workbench`.
-
+
2. If not already selected, click **Instances**, then **Instances**
3. Click **+ Create New**
diff --git a/images/10_chatbot_grounding.jpeg b/images/10_chatbot_grounding.jpeg
new file mode 100644
index 0000000..8f6826d
Binary files /dev/null and b/images/10_chatbot_grounding.jpeg differ
diff --git a/images/12_chatbot_grounding.jpeg b/images/12_chatbot_grounding.jpeg
new file mode 100644
index 0000000..9e1ff50
Binary files /dev/null and b/images/12_chatbot_grounding.jpeg differ
diff --git a/images/13_chatbot_grounding.jpeg b/images/13_chatbot_grounding.jpeg
new file mode 100644
index 0000000..b5dc1c4
Binary files /dev/null and b/images/13_chatbot_grounding.jpeg differ
diff --git a/images/14_chatbot_grounding.jpeg b/images/14_chatbot_grounding.jpeg
new file mode 100644
index 0000000..443e72d
Binary files /dev/null and b/images/14_chatbot_grounding.jpeg differ
diff --git a/images/1_select_vertexAI.jpeg b/images/1_select_vertexAI.jpeg
new file mode 100644
index 0000000..2124f7a
Binary files /dev/null and b/images/1_select_vertexAI.jpeg differ
diff --git a/images/7_chatbot_grounding.jpeg b/images/7_chatbot_grounding.jpeg
new file mode 100644
index 0000000..a56dbc8
Binary files /dev/null and b/images/7_chatbot_grounding.jpeg differ
diff --git a/images/8_chatbot_grounding.jpeg b/images/8_chatbot_grounding.jpeg
new file mode 100644
index 0000000..ce1d5f2
Binary files /dev/null and b/images/8_chatbot_grounding.jpeg differ
diff --git a/images/9_chatbot_grounding.jpeg b/images/9_chatbot_grounding.jpeg
new file mode 100644
index 0000000..3d7f76d
Binary files /dev/null and b/images/9_chatbot_grounding.jpeg differ
diff --git a/images/agent_builder1.jpeg b/images/agent_builder1.jpeg
new file mode 100644
index 0000000..2ddf606
Binary files /dev/null and b/images/agent_builder1.jpeg differ
diff --git a/images/agent_builder2.jpeg b/images/agent_builder2.jpeg
new file mode 100644
index 0000000..8bd5211
Binary files /dev/null and b/images/agent_builder2.jpeg differ
diff --git a/images/create-prompt.jpeg b/images/create-prompt.jpeg
new file mode 100644
index 0000000..62a7f68
Binary files /dev/null and b/images/create-prompt.jpeg differ
diff --git a/images/vertex-ai-create-prompt.jpeg b/images/vertex-ai-create-prompt.jpeg
new file mode 100644
index 0000000..926af76
Binary files /dev/null and b/images/vertex-ai-create-prompt.jpeg differ
diff --git a/notebooks/DL-gwas-gcp-example/1-d10-run-first.ipynb b/notebooks/DL-gwas-gcp-example/1-d10-run-first.ipynb
index 0f214ed..ce50864 100644
--- a/notebooks/DL-gwas-gcp-example/1-d10-run-first.ipynb
+++ b/notebooks/DL-gwas-gcp-example/1-d10-run-first.ipynb
@@ -34,7 +34,7 @@
"metadata": {},
"source": [
"## Prerequisites\n",
- "See the [README](/notebooks/DL-gwas-gcp-example/README.md) in this directory. "
+ "See the [README](./README.md) in this directory. "
]
},
{
diff --git a/notebooks/GWASCoatColor/GWAS_coat_color.ipynb b/notebooks/GWASCoatColor/GWAS_coat_color.ipynb
index c14ba4c..3cb22bd 100644
--- a/notebooks/GWASCoatColor/GWAS_coat_color.ipynb
+++ b/notebooks/GWASCoatColor/GWAS_coat_color.ipynb
@@ -81,7 +81,16 @@
"metadata": {},
"source": [
"#### Install dependencies\n",
- "Here we install mamba, which is faster than conda, but it can be tricky to add to path in a Sagemaker notebook so we just call the whole path. You could also skip this install and just use conda since that is preinstalled in the kernel."
+ "\n",
+ "The dependencies required to run this tutorial are `plink` and `vcftools`. A few different installation methods are given below. \n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "aa39cb82-ad03-4427-8173-2d5738e3eff5",
+ "metadata": {},
+ "source": [
+ "##### Method 1: Install with Mamba "
]
},
{
@@ -91,6 +100,7 @@
"metadata": {},
"outputs": [],
"source": [
+ "#If you don't have mamba installed, you can install it with the following command: \n",
"! curl -L -O https://github.com/conda-forge/miniforge/releases/latest/download/Mambaforge-$(uname)-$(uname -m).sh\n",
"! bash Mambaforge-$(uname)-$(uname -m).sh -b -p $HOME/mambaforge"
]
@@ -102,7 +112,7 @@
"metadata": {},
"outputs": [],
"source": [
- "#add to your path\n",
+ "#add mamba to your path\n",
"import os\n",
"os.environ[\"PATH\"] += os.pathsep + os.environ[\"HOME\"]+\"/mambaforge/bin\""
]
@@ -114,9 +124,52 @@
"metadata": {},
"outputs": [],
"source": [
+ "#Install dependencies with mamba\n",
"! mamba install -y -c bioconda plink vcftools"
]
},
+ {
+ "cell_type": "markdown",
+ "id": "720c6daa-4ee0-4db1-a94f-1f2b01e2f9f3",
+ "metadata": {},
+ "source": [
+ "##### Method 2: Install manually"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "1718f0a0-00b8-4d8b-8049-1bf705d1a8da",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#Plink installation \n",
+ "\n",
+ "# Please run these steps in the terminal\n",
+ "# 1. Download the PLINK 1.9 binary (Linux 64-bit)\n",
+ "! wget http://s3.amazonaws.com/plink1-assets/plink_linux_x86_64_20210606.zip\n",
+ "\n",
+ "# 2. Unzip the archive\n",
+ "! unzip plink_linux_x86_64_20210606.zip\n",
+ "\n",
+ "# 5. Move it to /usr/local/bin for global access (requires sudo)\n",
+ "! sudo mv plink /usr/local/bin/\n",
+ "\n",
+ "# 6. Check that it's installed correctly\n",
+ "! plink --version"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "891ccfbd-472d-4ebc-b326-6a81e19c2e6c",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# VCFTools Installation\n",
+ "! sudo apt-get -y install vcftools"
+ ]
+ },
{
"cell_type": "markdown",
"id": "3de2fc4c",
diff --git a/notebooks/GenAI/GCP_AI_Applications.ipynb b/notebooks/GenAI/GCP_AI_Applications.ipynb
index 3f0fd63..7746475 100644
--- a/notebooks/GenAI/GCP_AI_Applications.ipynb
+++ b/notebooks/GenAI/GCP_AI_Applications.ipynb
@@ -79,7 +79,7 @@
"metadata": {},
"source": [
"This tutorial will cost up to ~$6, pricing is based on the cost of AI Applications and Jupyter notebooks.\n",
- "For this tutorial we used a n2-standard-2 instance to run the python commands and created a App [Google Cloud Pricing Calculator](https://cloud.google.com/products/calculator?hl=en)."
+ "For this tutorial we used a n2-standard-2 instance to run the python commands and create an App. For more detailed information on how to calculate the price of various services on GCP, please access the [Google Cloud Pricing Calculator](https://cloud.google.com/products/calculator?hl=en)."
]
},
{
@@ -200,7 +200,7 @@
" doc_name=i.split(r'/')[-1]\n",
" x = requests.get(i)\n",
" doc = x.text\n",
- " upload_blob_from_memory(bucket, doc, {doc_name})"
+ " upload_blob_from_memory(bucket, doc, doc_name)"
]
},
{
@@ -220,7 +220,7 @@
"- Chatbots\n",
"- Search engine\n",
"- Recommendations bot\n",
- "- Agent development\n",
+ "- Conversational Agents\n",
"\n",
"For this tutorial we are creating a GenAI agent to help guide our chatbot in certain situations. Start by searching up **'AI Applications'** on the console. Then once you are on the page shown below click **'CREATE APP'**."
]
@@ -238,7 +238,7 @@
"id": "e6906e34-31a0-4e88-85e9-62abee5ed6cd",
"metadata": {},
"source": [
- "Click **'SELECT'** for the app named **Agent**."
+ "Click **'SELECT'** for the app named **Conversational Agent**."
]
},
{
@@ -246,7 +246,7 @@
"id": "39beb2ac-c377-4201-9973-5fdb738159d8",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
@@ -254,7 +254,7 @@
"id": "8d32f806-cef5-401b-be7b-b60ff2686a76",
"metadata": {},
"source": [
- "Give your agent a name, select your region, then click **'CREATE'**."
+ "Select **Build your own**. Give your agent a name, select your region, select **Playbook** and the conversation start method, then click **'CREATE'**."
]
},
{
@@ -262,7 +262,7 @@
"id": "0b59fa7c-b082-4372-9d9d-c299565f67dd",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
diff --git a/notebooks/GenAI/GCP_Code_Chatbot_wGrounding.ipynb b/notebooks/GenAI/GCP_Code_Chatbot_wGrounding.ipynb
index 9c63de6..91ba77a 100644
--- a/notebooks/GenAI/GCP_Code_Chatbot_wGrounding.ipynb
+++ b/notebooks/GenAI/GCP_Code_Chatbot_wGrounding.ipynb
@@ -11,9 +11,8 @@
"In this tutorial, we'll build a chatbot that utilizes data from a datastore that contains URLs relating to Snakemake information as its source. Grounding in this context means ensuring that the model's responses are strictly based on the information available on the website. The data store, which contains information from publicly indexed websites using a web crawler, allows you to specify domains and configure search or recommendation features based on the data collected from these sites. Grounding is a component of RAG but in this example, it operates at a higher level by the model assuming the information it already knows is not completely accurate. This ensures that the model fully depends on the data source for its responses. In turn, this helps RAG enhance the quality and accuracy of text generation by incorporating relevant information from external knowledge sources. For additional details on Agent Builder grounding, please refer to the __[GCP_Grounding](https://github.com/STRIDES/NIHCloudLabGCP/blob/main/notebooks/GenAI/GCP_Grounding.ipynb)__ tutorial. \n",
"\n",
"## Learning objectives\n",
- "- Learn to create a search app in Agent Builder.\n",
"- Learn to create a Website data store.\n",
- "- Learn to use an Agent Builder grounding.\n",
+ "- Learn to use an Vertex AI grounding.\n",
" \n",
"## Prerequisites\n",
"You must have enabled the Vertex AI, Compute Engine, and Agent Builder APIs.\n",
@@ -25,67 +24,7 @@
"Search LLM Add-On. Number of requests per month = 100
\n",
"Data Index. Amount of GiB indexed per month = 5
\n",
"\n",
- "## Get started\n",
- "\n",
- "### Create a search app\n",
- "\n",
- "As the __[GCP_Grounding](https://github.com/STRIDES/NIHCloudLabGCP/blob/main/notebooks/GenAI/GCP_Grounding.ipynb)__ tutorial explains, we need to create an Application under the 'Agent Builder' on the console. Start by searching up 'Agent Builder' on the console."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "f188bf06-2350-4baf-9e2a-915814288269",
- "metadata": {},
- "source": [
- ""
- ]
- },
- {
- "cell_type": "markdown",
- "id": "2a1588ce-2e85-4761-8137-98985fbc5fd1",
- "metadata": {},
- "source": [
- "Once you open Agent Builder click on 'CREATE APP' to get started."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "ce2cdbf4-c8dc-4020-b6b0-81238e1c10e0",
- "metadata": {},
- "source": [
- ""
- ]
- },
- {
- "cell_type": "markdown",
- "id": "dce20025-baf0-40f7-8d75-c5433f323237",
- "metadata": {},
- "source": [
- "Click on __\"SELECT\"__ to select an app Search type."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "30f25f07-3846-45d3-901b-981d18686a1d",
- "metadata": {},
- "source": [
- ""
- ]
- },
- {
- "cell_type": "markdown",
- "id": "c0757b89-94d7-4218-9a4f-99d6f5c8c031",
- "metadata": {},
- "source": [
- "The process of creating an application involves three steps: Type, Configuration, and Data. To set up the Search app configuration, choose the __\"Generic\"__ content option, enable both the __'Enterprise edition features'__ and __'Advanced LLM features'__, and then provide the application name along with the company or organization name."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "0123a946-7424-47b2-b9db-00b09e2f931c",
- "metadata": {},
- "source": [
- ""
+ "## Get started\n"
]
},
{
@@ -93,15 +32,7 @@
"id": "682ec6c3-7445-41ab-b75a-f76428568fdd",
"metadata": {},
"source": [
- "The next step is to create the data store. Click on __'CREATE DATA STORE'__ to proceed."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "763feba4-3e3a-4208-9896-5d82bf999b27",
- "metadata": {},
- "source": [
- ""
+ "The first next step is to create the data store. Click on __'CREATE DATA STORE'__ to proceed."
]
},
{
@@ -117,7 +48,7 @@
"id": "1dd57106-ccf0-488c-97fc-e773765884c9",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
@@ -125,15 +56,15 @@
"id": "051472e5-7a14-4d59-a26f-3d0d8068dd37",
"metadata": {},
"source": [
- "Selecting __'Website Content'__ will prompt you to enter a list of URLs and specify any sites to exclude. It is important to note that all the subdirectories of a domain can be extracted adding at the end of the URL a '__/*__'. i.e. in our case, we are extracting all the following webpages:\n",
+ "Selecting __'Website Content'__ will prompt you to enter a list of URLs and specify any sites to exclude. Enterinh https is not needed. It is important to note that all the subdirectories of a domain can be extracted adding at the end of the URL a '__/*__'. i.e. in our case, we are extracting all the following webpages:\n",
"```\n",
- "https://evodify.com/rna-seq-star-snakemake/*\n",
- "https://github.com/twbattaglia/RNAseq-workflow/*\n",
- "https://snakemake.readthedocs.io/en/stable/*\n",
- "https://snakemake.readthedocs.io/en/v4.5.0/*\n",
- "https://www.bioinformatics.babraham.ac.uk/training/Advanced_Python_Manual.docx\n",
- "https://www.cd-genomics.com/genomics.html/*\n",
- "https://www.cd-genomics.com/rna-seq-transcriptome.html/*\n",
+ "evodify.com/rna-seq-star-snakemake/*\n",
+ "github.com/twbattaglia/RNAseq-workflow/*\n",
+ "snakemake.readthedocs.io/en/stable/*\n",
+ "snakemake.readthedocs.io/en/v4.5.0/*\n",
+ "www.bioinformatics.babraham.ac.uk/training/Advanced_Python_Manual.docx\n",
+ "www.cd-genomics.com/genomics.html/*\n",
+ "www.cd-genomics.com/rna-seq-transcriptome.html/*\n",
"```\n",
"__Note:__ Make sure that 'Advanced website indexing' is __unchecked__. If the 'Advanced website indexing' option is enabled, you may encounter issues with grounding later on, as this option is intended for webpages owned by the user. For public webpages, ensure this option is disabled."
]
@@ -143,7 +74,7 @@
"id": "bb22e50f-fd47-46e4-b20a-5da96e096e03",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
@@ -159,7 +90,7 @@
"id": "933fae68-888d-40f4-b21f-281c7cee0bcc",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
@@ -175,7 +106,7 @@
"id": "adfe0682-195e-43e7-8418-165ea4de370d",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
@@ -183,7 +114,7 @@
"id": "9f692f25-8257-4e80-b3ea-06807ec83163",
"metadata": {},
"source": [
- "Once the application is created, the next step is to set up the chatbot. To do this, click on the three-line \"hamburger\" icon in the upper left corner of the window to view all available GCP products. Then, select __'Vertex AI'__ then under Vertex AI Studio select __'Chat'__. "
+ "Once the application is created, the next step is to set up the chatbot. To do this, click on the three-line \"hamburger\" icon in the upper left corner of the window to view all available GCP products. Then, select __'Vertex AI'__ then under Vertex AI Studio select __'Create Prompt'__. "
]
},
{
@@ -191,7 +122,7 @@
"id": "d7442aa8-c867-4716-b29e-7f9597b4b592",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
@@ -207,7 +138,7 @@
"id": "5007662b-149b-48e3-8b17-2bcc93f17638",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
@@ -223,7 +154,7 @@
"id": "49ca41d4-4e77-4175-8002-07732a10b92e",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
@@ -234,14 +165,6 @@
"Select __'Vertex AI search'__ as the grounding source, and in the __'Vertex AI datastore path'__ field, input the project ID, location, collections, and datastore ID in this format: __'projects/{PROJECT_ID}/locations/global/collections/default_collection/dataStores/{DATA_STORE_ID}'__. Once you've entered the required information, simply click save."
]
},
- {
- "cell_type": "markdown",
- "id": "456dee67-cd6a-4270-9191-017980d88d35",
- "metadata": {},
- "source": [
- ""
- ]
- },
{
"cell_type": "markdown",
"id": "124b324f-f513-447c-8f0f-fba69fbb20e4",
@@ -318,7 +241,7 @@
"source": [
"## Conclusion\n",
"\n",
- "You have learned how to create a Website data store using the Agent Builder and Grounding in Vertex AI Studio. Additionally, you now understand the key parameters needed to properly configure grounding for accurate information extraction from the Website data store."
+ "You have learned how to create a Website data store using the AI Applications and Grounding in Vertex AI Studio. Additionally, you now understand the key parameters needed to properly configure grounding for accurate information extraction from the Website data store."
]
},
{
@@ -330,14 +253,6 @@
"\n",
"Please remember to delete or stop your Jupyter notebook and delete your data store and search app on Agent Builder to prevent incurring charges. And if you have created any other services like buckets, please remember to delete them as well."
]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "757a159b-5550-4965-a249-0e16753a9b76",
- "metadata": {},
- "outputs": [],
- "source": []
}
],
"metadata": {},
diff --git a/notebooks/GenAI/GCP_Grounding.ipynb b/notebooks/GenAI/GCP_Grounding.ipynb
index 2f8b6a5..cd4c11d 100644
--- a/notebooks/GenAI/GCP_Grounding.ipynb
+++ b/notebooks/GenAI/GCP_Grounding.ipynb
@@ -21,8 +21,7 @@
"id": "fbb045c4-b33f-4499-8d3d-c8545206e3ac",
"metadata": {},
"source": [
- "Google Cloud's [Grounding](https://cloud.google.com/vertex-ai/generative-ai/docs/grounding/overview) feature is designed to improve the accuracy and relevance of search results by understanding the context of your search query and the information presented on web pages or your own data via the console. This requires a search app and a data store which both will be created in another Google Cloud product called [Agent Builder\n",
- "](https://cloud.google.com/products/agent-builder?hl=en)."
+ "Google Cloud's [Grounding](https://cloud.google.com/vertex-ai/generative-ai/docs/grounding/overview) feature is designed to improve the accuracy and relevance of search results by understanding the context of your search query and the information presented on web pages or your own data via the console. a data store which will be created in another Google Cloud product called [AI Applications](https://cloud.google.com/products/agent-builder?hl=en)."
]
},
{
@@ -66,64 +65,6 @@
"## Get started"
]
},
- {
- "cell_type": "markdown",
- "id": "ed6e13da-8475-4fb5-9090-e68638d761f5",
- "metadata": {},
- "source": [
- "### Create a search app"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "bfe28a30-ec48-4257-b342-1fc8c3954510",
- "metadata": {},
- "source": [
- "In order to utilize grounding with your own data you first need to create a search app. Start by searching up **'Agent Builder'** on the console. Then once you are on the page shown below click **'CREATE APP'**."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "9407c375-7b45-40b6-8504-3d6eced555da",
- "metadata": {},
- "source": [
- ""
- ]
- },
- {
- "cell_type": "markdown",
- "id": "e6906e34-31a0-4e88-85e9-62abee5ed6cd",
- "metadata": {},
- "source": [
- "Click **'SELECT'** for the app named **Search**."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "39beb2ac-c377-4201-9973-5fdb738159d8",
- "metadata": {},
- "source": [
- ""
- ]
- },
- {
- "cell_type": "markdown",
- "id": "20abe50b-ebf4-4214-a494-b199ac7e56bb",
- "metadata": {},
- "source": [
- "You have the option to tailor what kind of search you would like to use. For this tutorial we will be using **'Generic'**. The other two options include searching through media files and Healthcare files in FHIR format.\n",
- "\n",
- "Also make sure that you have enabled the **'Enterprise edition feature'** and **'Advance LLM feature'** to allow your app to be searchable via the grounding feature and if prompted set your location to **'global'**."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "d2d4afa0-bdf7-4afc-8b74-d3e791bbaebd",
- "metadata": {},
- "source": [
- ""
- ]
- },
{
"cell_type": "markdown",
"id": "6b89e7d4-bbf8-4ce9-890e-4ff9f17b4610",
@@ -137,7 +78,7 @@
"id": "2322c0da-1554-47c9-9e27-2732beda35dd",
"metadata": {},
"source": [
- "Next we'll create a data store that will allow us to connect a data source to our search app."
+ "First we'll create a data store that will allow us to connect a specific data source. Navigate to `AI Applications` > `Data Stores` > `Create Data Store`. "
]
},
{
@@ -153,7 +94,7 @@
"id": "ccd0d718-eb6f-43b3-afe1-ec56d8108ef3",
"metadata": {},
"source": [
- "As you can see from the image below Google Cloud's Agent builder supports the following data sources but for this tutorial we will use **'Cloud Storage'** as our data source."
+ "As you can see from the image below Google Cloud's AI Applications supports the following data sources but for this tutorial we will use **'Cloud Storage'** as our data source."
]
},
{
@@ -185,7 +126,7 @@
"id": "332994e1-1bb8-476e-bbb0-28a1169e86b7",
"metadata": {},
"source": [
- "Configure your data store by giving it a name and make sure you have set your location to **global**."
+ "Configure your data store by giving it a name and make sure you have set your location to **global**. And Select `Create`. "
]
},
{
@@ -196,36 +137,12 @@
""
]
},
- {
- "cell_type": "markdown",
- "id": "ebeba3cc-72dd-4758-8584-96214f9ade6a",
- "metadata": {},
- "source": [
- "### Sync your data"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "51310b87-7a49-4211-bd91-fe67c45274f4",
- "metadata": {},
- "source": [
- "Select your data store or multiple data stores and click create to sync them to your search app. "
- ]
- },
- {
- "cell_type": "markdown",
- "id": "c0f2e40e-320a-4fd9-8795-d273f00557d3",
- "metadata": {},
- "source": [
- ""
- ]
- },
{
"cell_type": "markdown",
"id": "d4002baa-ae55-492f-b9d8-e9729a872db8",
"metadata": {},
"source": [
- "Depending on the size of your data it can take roughly ~30min to sync the date to your app but once its ready you should see a green check mark by the date of the last document import date similar to the image below. Make sure you **copy the data store ID** so we can use it later."
+ "Make sure you **copy the data store ID** so we can use it later."
]
},
{
@@ -249,7 +166,7 @@
"id": "369c112d-2376-43fa-9552-9498479be9fd",
"metadata": {},
"source": [
- "On the console go to Vertex AI and head down to Vertex AI Studio where the playground is. Select the **Chat** playground. "
+ "On the console go to Vertex AI and head down to Vertex AI and Click on Create Prompt. "
]
},
{
@@ -257,7 +174,7 @@
"id": "8c971978-4925-427c-900c-0e8a996c0c91",
"metadata": {},
"source": [
- ""
+ ""
]
},
{
@@ -265,7 +182,7 @@
"id": "5566af32-8ae4-42e2-8a03-cb2af69a6e60",
"metadata": {},
"source": [
- "On the right side menu select **'Enable Grounding'** then click **'CUSTOMIZE'**."
+ "On the right side menu select **'Enable Grounding'** then click **'CUSTOMIZE'**. Select **Vertex AI Search** as the grounding source. "
]
},
{
diff --git a/notebooks/GenAI/GCP_RAG_for_Structure_Data.ipynb b/notebooks/GenAI/GCP_RAG_for_Structure_Data.ipynb
index 9146c01..26b68ac 100644
--- a/notebooks/GenAI/GCP_RAG_for_Structure_Data.ipynb
+++ b/notebooks/GenAI/GCP_RAG_for_Structure_Data.ipynb
@@ -126,7 +126,7 @@
"id": "64e22773-9a03-47de-9b6d-7946abcba722",
"metadata": {},
"source": [
- "Set your project id, location, and bucket variables."
+ "Set your project id, location, and bucket variables. Ensure that the dataset name only contains alphanumeric characters and underscores. "
]
},
{
@@ -136,11 +136,11 @@
"metadata": {},
"outputs": [],
"source": [
- "project_id=''\n",
- "location=' (e.g.us-east4)'\n",
- "bucket = ''\n",
- "dataset_name = ''\n",
- "table_name = \"\""
+ "project_id=''\n",
+ "location=''\n",
+ "bucket = ''\n",
+ "dataset_name = ''\n",
+ "table_name = \"\""
]
},
{
@@ -169,7 +169,7 @@
"source": [
"Once the bucket is created, we need to access the CSV source file. In this tutorial, we transferred the data file to our Jupyter notebook by simply dragging and dropping it from my local folder. Next, we need to specify the bucket name and the path of the data source in order to upload the CSV file to the bucket. \n",
"\n",
- "We are using the [health screening](https://www.kaggle.com/datasets/drateendrajha/health-screening-data) dataset from kaggle for this tutorial."
+ "We are using the [Data Science Salaries](https://www.kaggle.com/datasets/ajjarvis/ds-salaries) dataset from kaggle for this tutorial."
]
},
{
@@ -179,7 +179,7 @@
"metadata": {},
"outputs": [],
"source": [
- "!gsutil cp '' gs://{bucket}"
+ "!gsutil cp 'ds_salaries.csv' gs://{bucket}"
]
},
{
@@ -430,7 +430,7 @@
"source": [
"query = f\"\"\"\n",
" SELECT count(gender) as cgender\n",
- " FROM `{project_id.dataset_id.table_id}`\n",
+ " FROM '{project_id.dataset_id.table_id}'\n",
"\"\"\"\n",
"\n",
"# Execute the query\n",
@@ -452,15 +452,6 @@
" print(f\"Error executing query: {str(e)}\")"
]
},
- {
- "cell_type": "markdown",
- "id": "ce65ead3-694d-4d46-9376-38d35b8d0852",
- "metadata": {},
- "source": [
- "cgender: 139920\n",
- ""
- ]
- },
{
"cell_type": "markdown",
"id": "b0cd8a0d-9845-4d84-abf6-2610ed946b7e",
@@ -519,7 +510,6 @@
" model=\"gemini-1.5-pro\",\n",
" temperature=0,\n",
" max_tokens=8190,\n",
- " timeout=None,\n",
" max_retries=2,\n",
")\n",
"db = SQLDatabase.from_uri(sqlalchemy_url)\n",
diff --git a/notebooks/GenAI/Gemini_Intro.ipynb b/notebooks/GenAI/Gemini_Intro.ipynb
index c6a24d8..5b2f106 100644
--- a/notebooks/GenAI/Gemini_Intro.ipynb
+++ b/notebooks/GenAI/Gemini_Intro.ipynb
@@ -93,8 +93,8 @@
"from vertexai.generative_models import GenerativeModel, Image, GenerativeModel, ChatSession, Part, GenerationConfig\n",
"\n",
"# TODO(developer): Update and un-comment below lines\n",
- "project_id = \"\"\n",
- "location = \"\" #(e.g., us-central1)\n",
+ "project_id = \"\"\n",
+ "location = \"\" #(e.g., us-central1)\n",
"vertexai.init(project=project_id, location=location)"
]
},
@@ -129,7 +129,7 @@
"metadata": {},
"outputs": [],
"source": [
- "model = GenerativeModel(\"gemini-pro\")\n",
+ "model = GenerativeModel(\"gemini-2.0-flash-001\")\n",
"chat = model.start_chat()\n",
"\n",
"def get_chat_response(chat: ChatSession, prompt: str):\n",
@@ -211,14 +211,11 @@
]
},
{
- "cell_type": "code",
- "execution_count": null,
- "id": "ee4cf184-c815-425c-9742-7625123e02bf",
+ "cell_type": "markdown",
+ "id": "66a897a5-79e7-4705-8ef9-22f0807cbca8",
"metadata": {},
- "outputs": [],
"source": [
- "#download the article\n",
- "!wget --user-agent \"Chrome\" https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10954554/pdf/41586_2024_Article_7159.pdf"
+ "Download the PDF document we will be using for summarization from the following [link](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10954554/pdf/41586_2024_Article_7159.pdf). Then upload the document to your working folder."
]
},
{
@@ -330,7 +327,7 @@
"outputs": [],
"source": [
"def img2text(image_path: str, img_prompt: str) -> str:\n",
- " multimodal_model = GenerativeModel(\"gemini-pro-vision\")\n",
+ " multimodal_model = GenerativeModel(\"gemini-2.0-flash\")\n",
" if \"gs://\" in image_path:\n",
" image1=Part.from_uri(image_path, mime_type=\"image/jpeg\")\n",
" else: \n",
@@ -465,7 +462,7 @@
"source": [
"def video2text(video_path: str, video_prompt: str) -> str:\n",
" # Query the model\n",
- " multimodal_model = GenerativeModel(\"gemini-pro-vision\")\n",
+ " multimodal_model = GenerativeModel(\"gemini-2.0-flash\")\n",
" response = multimodal_model.generate_content(\n",
" [\n",
" # Add an example image\n",
diff --git a/notebooks/GenAI/Google_Drive_chatbot.ipynb b/notebooks/GenAI/Google_Drive_chatbot.ipynb
index 6ae70ec..90560aa 100644
--- a/notebooks/GenAI/Google_Drive_chatbot.ipynb
+++ b/notebooks/GenAI/Google_Drive_chatbot.ipynb
@@ -53,7 +53,7 @@
"source": [
"We assume you have access to **Vertex AI** and Google Drive and have enabled the APIs. If not go to the console then `APIs & Services > Enabled APIs & Services` search for **'Google Drive API'** then click 'Enable'. do the same for Vertex AI.\n",
"\n",
- "In this tutorial we will be using Google **Gemini Pro 1.5** which doesn't need to be deployed but if you would like to use another model you choose one from the **Model Garden** using the console which will allow you to add a model to your model registry, create an endpoint (or use an existing one), and deploy the model all in one step. \n",
+ "In this tutorial we will be using Google **gemini-2.0-flash** which doesn't need to be deployed but if you would like to use another model you choose one from the **Model Garden** using the console which will allow you to add a model to your model registry, create an endpoint (or use an existing one), and deploy the model all in one step. \n",
"\n",
"The last thing before we begin will to create a **Vertex AI RAG Data Service Agent** service account by going to `IAM` on the console then check mark **Include Google-provided role grant** if it not listed there then click grant access and add Vertex AI RAG Data Service Agent as a role."
]
@@ -134,9 +134,9 @@
"metadata": {},
"outputs": [],
"source": [
- "project_id = \"\"\n",
- "display_name = \"\"\n",
- "location = \" (e.g., us-central1)"
+ "project_id = \"\"\n",
+ "display_name = \"\"\n",
+ "location = \"\"#Please ensure that you are using a region that supports the creation of a RAG Corpus (e.g. us-central1)"
]
},
{
@@ -378,7 +378,7 @@
"id": "a3a122f7-0734-45e2-b93c-c93eebfbe45b",
"metadata": {},
"source": [
- "Now we can create a RAG retrieval tool that will allow us to connect our model to 1 corpus to retrieval relevant data from. Notice we are using **gemini-1.0-pro-002**."
+ "Now we can create a RAG retrieval tool that will allow us to connect our model to 1 corpus to retrieval relevant data from. Notice we are using **gemini-2.0-flash**."
]
},
{
@@ -400,7 +400,7 @@
")\n",
"# Create a gemini-pro model instance\n",
"rag_model = GenerativeModel(\n",
- " model_name=\"gemini-1.0-pro-002\", tools=[rag_retrieval_tool]\n",
+ " model_name=\"gemini-2.0-flash\", tools=[rag_retrieval_tool]\n",
")\n"
]
},
@@ -473,7 +473,7 @@
" )\n",
" # Create a gemini-pro model instance\n",
" rag_model = GenerativeModel(\n",
- " model_name=\"gemini-1.0-pro-002\", tools=[rag_retrieval_tool]\n",
+ " model_name=\"gemini-2.0-flash\", tools=[rag_retrieval_tool]\n",
" )\n",
" response = rag_model.generate_content(question)\n",
" \n",
@@ -542,6 +542,14 @@
"source": [
"If you have imported a model and deployed it don't forget to delete the model from the Model Registry and delete the endpoint."
]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "c61a78f9-6e8b-4897-90ce-54e6268773f4",
+ "metadata": {},
+ "outputs": [],
+ "source": []
}
],
"metadata": {},
diff --git a/notebooks/GenAI/Pubmed_RAG_chatbot.ipynb b/notebooks/GenAI/Pubmed_RAG_chatbot.ipynb
index 9d4bcf5..5f2719c 100644
--- a/notebooks/GenAI/Pubmed_RAG_chatbot.ipynb
+++ b/notebooks/GenAI/Pubmed_RAG_chatbot.ipynb
@@ -21,7 +21,7 @@
"id": "3ecea2ad-7c65-4367-87e1-b021167c3a1d",
"metadata": {},
"source": [
- "For this tutorial we create a PubMed chatbot that will answer questions by gathering information from documents we have provided via an index. The model we will be using today is the pretrained 'test-bison@001' model from GCP."
+ "For this tutorial we create a PubMed chatbot that will answer questions by gathering information from documents we have provided via an index. The model we will be using today is the pretrained 'gemini-2.0-flash' model from GCP."
]
},
{
@@ -65,7 +65,7 @@
"id": "9dbd13e7-afc9-416b-94dc-418a93e14587",
"metadata": {},
"source": [
- "In this tutorial we will be using Google PaLM2 LLM **test-bison@001** which doesn't need to be deployed but if you would like to use another model you choose one from the **Model Garden** using the console which will allow you to add a model to your model registry, create an endpoint (or use an existing one), and deploy the model all in one step."
+ "In this tutorial we will be using a Google's gemini model **gemini-2.0-flash** which doesn't need to be deployed but if you would like to use another model you choose one from the **Model Garden** using the console which will allow you to add a model to your model registry, create an endpoint (or use an existing one), and deploy the model all in one step."
]
},
{
@@ -133,9 +133,9 @@
"metadata": {},
"outputs": [],
"source": [
- "project_id=''\n",
- "location=' (e.g.us-east4)'\n",
- "bucket = ''"
+ "project_id='PROJECT_ID'\n",
+ "location='REGION'\n",
+ "bucket = 'UNIQUE_BUCKET_NAME'"
]
},
{
@@ -589,7 +589,7 @@
"source": [
"from langchain_google_vertexai import VectorSearchVectorStore\n",
"from langchain_google_vertexai import VertexAIEmbeddings\n",
- "embeddings = VertexAIEmbeddings(model_name=\"textembedding-gecko@003\")\n",
+ "embeddings = VertexAIEmbeddings(model_name=\"text-embedding-005\")\n",
"\n",
"# initialize vector store\n",
"vector_store = VectorSearchVectorStore.from_components(\n",
@@ -710,7 +710,7 @@
"- **ConversationalRetrievalChain:** Allows the user to construct a conversation with the model and retrieves the outputs while sending inputs to the model.\n",
"- **PromptTemplate:** Allows the user to prompt the model to provide instructions, best method for zero and few shot prompting\n",
"- **VertexAIEmbeddings:** Text embedding model used before to convert text to numerical vectors.\n",
- "- **VertexAI**: Package used to import Google PaLM2 LLMs models (e.g. text-bison@001, code-bison). \n"
+ "- **VertexAI**: Package used to import Google PaLM2 LLMs models. \n"
]
},
{
@@ -793,7 +793,7 @@
"id": "dab1012f-ed20-47b9-9162-924e03e836d5",
"metadata": {},
"source": [
- "Now we can define our Google PaLM2 model being `text-bison@001` and other parameters:\n",
+ "Now we can define our Google PaLM2 model being `gemini-2.0-flash` and other parameters:\n",
"\n",
"- Max Output Tokens: Limit of tokens outputted by the model.\n",
"- Temperature: Controls randomness, higher values increase diversity meaning a more unique response make the model to think harder. Must be a number from 0 to 1, 0 being less unique.\n",
@@ -808,7 +808,7 @@
"source": [
"```python\n",
"llm = VertexAI(\n",
- " model_name=\"chat-bison@002\",\n",
+ " model_name=\"gemini-2.0-flash\",\n",
" max_output_tokens=1024,\n",
" temperature=0.2,\n",
" top_p=0.8,\n",
@@ -841,7 +841,7 @@
"\n",
"#only if using Vector Search as a retriever\n",
"\n",
- "embeddings = VertexAIEmbeddings(model_name=\"textembedding-gecko@003\") #Make sure embedding model is compatible with model\n",
+ "embeddings = VertexAIEmbeddings(model_name=\"text-embedding-005\") #Make sure embedding model is compatible with model\n",
"\n",
"vector_store = VectorSearchVectorStore.from_components(\n",
" project_id=PROJECT_ID,\n",
diff --git a/notebooks/GenAI/VertexAIStudioGCP.ipynb b/notebooks/GenAI/VertexAIStudioGCP.ipynb
index d217321..5bd4b92 100644
--- a/notebooks/GenAI/VertexAIStudioGCP.ipynb
+++ b/notebooks/GenAI/VertexAIStudioGCP.ipynb
@@ -63,19 +63,24 @@
"cell_type": "markdown",
"metadata": {},
"source": [
- "Go to the Vertex AI Studio console by navigating to Vertex AI via the search bar on the console. On the left side menu scroll down to Vertex AI Studio, click **Chat**.\n",
+ "Click on the horizontal three lines on the top left of the console, scroll down to **Vertex AI**, click **Prompt Gallery**. \n",
"\n",
- "  \n",
+ "
\n",
+ "  \n",
"\n",
"\n",
- "Click on **\"Browse prompt gallery\"** then scroll down to **Summarizing texts** and click on it. You will see the system instructions and the prompt on the chat that are populated with the summarizing example. Since we want to summarize our own article, we clear the prompt by clicking **Clear prompt** on the top right and then **Insert Media**, select **ByURL** if you want to upload a document from the web or **Upload** if you want to upload it from your local machine. Paste the url link and press **Insert**. The document will be loaded into the Prompt and you will have to enter some instruction of what to do with the uploaded document i.e. \"Please summarize the uploaded document.\" For this tutorial this article is about how gut microbiota affects Alzeheimer's disease because of the gut-brain-microbiota axis network [here](https://www.aging-us.com/article/102930/pdf).\n",
- "\n",
+ "Search for **Summarizing texts** and click on it. You will see the system instructions and the prompt on the chat that are populated with the summarizing example. Since we want to summarize our own article, we clear the prompt by clicking **Clear prompt** on the top right and then **Insert Media**, select **ByURL** if you want to upload a document from the web or **Upload** if you want to upload it from your local machine. Paste the url link and press **Insert**. The document will be loaded into the Prompt and you will have to enter some instruction of what to do with the uploaded document i.e. \"Please summarize the uploaded document.\" For this tutorial this article is about how gut microbiota affects Alzeheimer's disease because of the gut-brain-microbiota axis network [here](https://www.aging-us.com/article/102930/pdf).\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
"
\n",
"\n",
"\n",
- "Click on **\"Browse prompt gallery\"** then scroll down to **Summarizing texts** and click on it. You will see the system instructions and the prompt on the chat that are populated with the summarizing example. Since we want to summarize our own article, we clear the prompt by clicking **Clear prompt** on the top right and then **Insert Media**, select **ByURL** if you want to upload a document from the web or **Upload** if you want to upload it from your local machine. Paste the url link and press **Insert**. The document will be loaded into the Prompt and you will have to enter some instruction of what to do with the uploaded document i.e. \"Please summarize the uploaded document.\" For this tutorial this article is about how gut microbiota affects Alzeheimer's disease because of the gut-brain-microbiota axis network [here](https://www.aging-us.com/article/102930/pdf).\n",
- "\n",
+ "Search for **Summarizing texts** and click on it. You will see the system instructions and the prompt on the chat that are populated with the summarizing example. Since we want to summarize our own article, we clear the prompt by clicking **Clear prompt** on the top right and then **Insert Media**, select **ByURL** if you want to upload a document from the web or **Upload** if you want to upload it from your local machine. Paste the url link and press **Insert**. The document will be loaded into the Prompt and you will have to enter some instruction of what to do with the uploaded document i.e. \"Please summarize the uploaded document.\" For this tutorial this article is about how gut microbiota affects Alzeheimer's disease because of the gut-brain-microbiota axis network [here](https://www.aging-us.com/article/102930/pdf).\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
"  \n",
"\n",
"To the right you can control the parameters that we have been using before this is a great way to test what each parameter does and how they effect each other. Once you are done click the submit arrow, you should have a similar output as below. For explainations on the parameters **temperature, Output token limit, top p, and top k** see the following article [here](https://cloud.google.com/vertex-ai/docs/generative-ai/text/test-text-prompts#generative-ai-test-text-prompt-drest).\n",
"\n",
"
\n",
"\n",
"To the right you can control the parameters that we have been using before this is a great way to test what each parameter does and how they effect each other. Once you are done click the submit arrow, you should have a similar output as below. For explainations on the parameters **temperature, Output token limit, top p, and top k** see the following article [here](https://cloud.google.com/vertex-ai/docs/generative-ai/text/test-text-prompts#generative-ai-test-text-prompt-drest).\n",
"\n",
"  \n",
- " \n"
+ " "
]
},
{
diff --git a/notebooks/GenAI/example_scripts/example_langchain_chat_llama_2_zeroshot.py b/notebooks/GenAI/example_scripts/example_langchain_chat_llama_2_zeroshot.py
index e554caf..27c86cf 100644
--- a/notebooks/GenAI/example_scripts/example_langchain_chat_llama_2_zeroshot.py
+++ b/notebooks/GenAI/example_scripts/example_langchain_chat_llama_2_zeroshot.py
@@ -3,6 +3,7 @@
from langchain.prompts import PromptTemplate
#from langchain.llms import VertexAIModelGarden
from langchain_google_vertexai import ChatVertexAI
+from langchain_google_vertexai import VertexAI
import sys
import json
import os
@@ -30,7 +31,7 @@ def build_chain():
#llm = VertexAIModelGarden(project=PROJECT_ID, endpoint_id=ENDPOINT_ID, location=LOCATION_ID)
llm = VertexAI(
- model_name="chat-bison@002",
+ model_name="gemini-2.0-flash",
max_output_tokens=1024,
temperature=0.2,
top_p=0.8,
diff --git a/notebooks/GenAI/example_scripts/example_vectorsearch_chat_llama_2_zeroshot.py b/notebooks/GenAI/example_scripts/example_vectorsearch_chat_llama_2_zeroshot.py
index 85a9033..fef58ca 100644
--- a/notebooks/GenAI/example_scripts/example_vectorsearch_chat_llama_2_zeroshot.py
+++ b/notebooks/GenAI/example_scripts/example_vectorsearch_chat_llama_2_zeroshot.py
@@ -4,6 +4,7 @@
from langchain_google_vertexai import VertexAIEmbeddings
from langchain_google_vertexai import VectorSearchVectorStore
from langchain_google_vertexai import ChatVertexAI
+from langchain_google_vertexai import VertexAI
import sys
import json
import os
@@ -35,7 +36,7 @@ def build_chain():
#llm = VertexAIModelGarden(project=PROJECT_ID, endpoint_id=ENDPOINT_ID, location=LOCATION_ID)
llm = VertexAI(
- model_name="chat-bison@002",
+ model_name="gemini-2.0-flash",
max_output_tokens=1024,
temperature=0.2,
top_p=0.8,
diff --git a/notebooks/GenAI/langchain_on_vertex.ipynb b/notebooks/GenAI/langchain_on_vertex.ipynb
index 105e3ec..5dcee40 100644
--- a/notebooks/GenAI/langchain_on_vertex.ipynb
+++ b/notebooks/GenAI/langchain_on_vertex.ipynb
@@ -89,7 +89,9 @@
"from langchain_community.document_loaders import WebBaseLoader\n",
"from langchain.chains.summarize import load_summarize_chain\n",
"from langchain.schema.prompt_template import format_document\n",
- "from langchain_text_splitters import RecursiveCharacterTextSplitter"
+ "from langchain_text_splitters import RecursiveCharacterTextSplitter\n",
+ "\n",
+ "! export USER_AGENT=\"my-langchain-script/1.0\""
]
},
{
@@ -108,7 +110,7 @@
"outputs": [],
"source": [
"loader = WebBaseLoader(\"https://pubmed.ncbi.nlm.nih.gov/37883540/\")\n",
- "docs = loader.load()"
+ "docs = loader.load()\n"
]
},
{
@@ -118,7 +120,7 @@
"metadata": {},
"outputs": [],
"source": [
- "llm = ChatVertexAI()\n",
+ "llm = ChatVertexAI(model_name=\"gemini-1.5-flash-002\")\n",
"print('the LLM and default params are : ', llm)\n",
"\n",
"chain = load_summarize_chain(llm, chain_type=\"stuff\")\n",
@@ -274,7 +276,7 @@
"outputs": [],
"source": [
"# index the document using FAISS\n",
- "embeddings = VertexAIEmbeddings(model_name=\"textembedding-gecko@003\")\n",
+ "embeddings = VertexAIEmbeddings(model_name=\"text-embedding-004\")\n",
"faiss_index = FAISS.from_documents(pages, embeddings)"
]
},
@@ -331,7 +333,7 @@
" )\n",
" }\n",
" | PromptTemplate.from_template(\"Summarize the following content in around 200 words:\\n\\n{content}\")\n",
- " | ChatVertexAI()\n",
+ " | ChatVertexAI(model=\"gemini-1.5-flash-002\")\n",
" | StrOutputParser()\n",
")"
]
@@ -388,7 +390,7 @@
" )\n",
" }\n",
" | PromptTemplate.from_template(prompt_str) \n",
- " | ChatVertexAI()\n",
+ " | ChatVertexAI(model=\"gemini-1.5-flash-002\")\n",
" | StrOutputParser()\n",
")"
]
@@ -483,7 +485,7 @@
"metadata": {},
"outputs": [],
"source": [
- "llm = ChatVertexAI(model_name=\"codechat-bison@002\", max_output_tokens=1000, temperature=0.3)"
+ "llm = ChatVertexAI(model_name=\"gemini-2.0-flash-001\", max_output_tokens=1000, temperature=0.3)"
]
},
{
diff --git a/notebooks/GoogleBatch/nextflow/Part1_GBatch_Nextflow.ipynb b/notebooks/GoogleBatch/nextflow/Part1_GBatch_Nextflow.ipynb
index 9b61d1c..717365d 100644
--- a/notebooks/GoogleBatch/nextflow/Part1_GBatch_Nextflow.ipynb
+++ b/notebooks/GoogleBatch/nextflow/Part1_GBatch_Nextflow.ipynb
@@ -187,7 +187,7 @@
" ]\n",
" }\n",
" }\n",
- " ]\n",
+ " ],\n",
" \"volumes\": [\n",
" {\n",
" \"gcs\": {\n",
diff --git a/notebooks/GoogleBatch/nextflow/Part2_GBatch_Nextflow.ipynb b/notebooks/GoogleBatch/nextflow/Part2_GBatch_Nextflow.ipynb
index 1e0e8fe..debf12d 100644
--- a/notebooks/GoogleBatch/nextflow/Part2_GBatch_Nextflow.ipynb
+++ b/notebooks/GoogleBatch/nextflow/Part2_GBatch_Nextflow.ipynb
@@ -180,9 +180,26 @@
"outputs": [],
"source": [
"#Install SRAtools to download data\n",
+ "\n",
+ "#Option 1: Using mamba\n",
"! mamba install -c bioconda -c conda-forge sra-tools==2.11.0 -y"
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "1242056b-39f3-435d-b0d4-2757c3faa0ff",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#Option 2: Manual Install with wget \n",
+ "\n",
+ "!wget https://ftp-trace.ncbi.nlm.nih.gov/sra/sdk/current/sratoolkit.current-ubuntu64.tar.gz && \\\n",
+ "tar -xvzf sratoolkit.current-ubuntu64.tar.gz && \\\n",
+ "export PATH=$PATH:$(pwd)/sratoolkit.*-ubuntu64/bin && \\\n",
+ "fasterq-dump --version"
+ ]
+ },
{
"cell_type": "markdown",
"id": "1bc477a9-7b3f-431e-93b7-50e96809bfc5",
@@ -238,7 +255,7 @@
"outputs": [],
"source": [
"%%time\n",
- "! fasterq-dump -f -e 8 -m 24G SRR067701.sra"
+ "! fasterq-dump -f -e 8 -m 24G SRR067701"
]
},
{
diff --git a/notebooks/SRADownload/SRA-Download.ipynb b/notebooks/SRADownload/SRA-Download.ipynb
index 665d19b..30171a8 100644
--- a/notebooks/SRADownload/SRA-Download.ipynb
+++ b/notebooks/SRADownload/SRA-Download.ipynb
@@ -59,9 +59,17 @@
"id": "01213dae",
"metadata": {},
"source": [
- "Install dependencies, using mamba (you could also use conda). At the time of writing, the version of SRA tools available with the Anaconda distribution was v.2.11.0. If you want to install the latest version, download and install from [here](https://github.com/ncbi/sra-tools/wiki/01.-Downloading-SRA-Toolkit). If you do the direct install, you will also need to configure interactively following [this guide](https://github.com/ncbi/sra-tools/wiki/05.-Toolkit-Configuration), you can do that by opening a terminal and running the commands there."
+ "Install dependencies, using mamba (you could also use conda) or manually by downloading and installing the package. At the time of writing, the version of SRA tools available with the Anaconda distribution was v.2.11.0. If you want to install the latest version, download and install from [here](https://github.com/ncbi/sra-tools/wiki/01.-Downloading-SRA-Toolkit). If you do the direct install, you will also need to configure interactively following [this guide](https://github.com/ncbi/sra-tools/wiki/05.-Toolkit-Configuration), you can do that by opening a terminal and running the commands there."
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "3e0bb6f1-05ee-4215-94e3-8365f7550688",
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ },
{
"cell_type": "code",
"execution_count": null,
@@ -90,6 +98,19 @@
"! fasterq-dump -h"
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "5df3de21-bb3d-4d38-bcb1-30f5b10c9d01",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "!wget https://ftp-trace.ncbi.nlm.nih.gov/sra/sdk/current/sratoolkit.current-ubuntu64.tar.gz && \\\n",
+ "tar -xvzf sratoolkit.current-ubuntu64.tar.gz && \\\n",
+ "export PATH=$PATH:$(pwd)/sratoolkit.*-ubuntu64/bin && \\\n",
+ "fasterq-dump --version"
+ ]
+ },
{
"cell_type": "markdown",
"id": "7d8d3512-5307-42f7-9405-495fe1ca5be2",
diff --git a/notebooks/SpleenLiverSegmentation/README.md b/notebooks/SpleenLiverSegmentation/README.md
index 976552f..e480f8b 100644
--- a/notebooks/SpleenLiverSegmentation/README.md
+++ b/notebooks/SpleenLiverSegmentation/README.md
@@ -9,8 +9,7 @@ The Spleen model is additionally retrained on the medical decathlon spleen datas
Data is not necessary to be downloaded to run the notebook. The notebook downloads the data during it's run.
The notebook uses the Python package [MONAI](https://monai.io/), the Medical Open Network for Artificial Intelligence.
-- Spleen Model - [clara_pt_spleen_ct_segmentation_V2](https://catalog.ngc.nvidia.com/orgs/nvidia/teams/monaitoolkit/models/monai_spleen_ct_segmentation)
-- Liver Model - [clara_pt_liver_and_tumor_ct_segmentation_V1]()
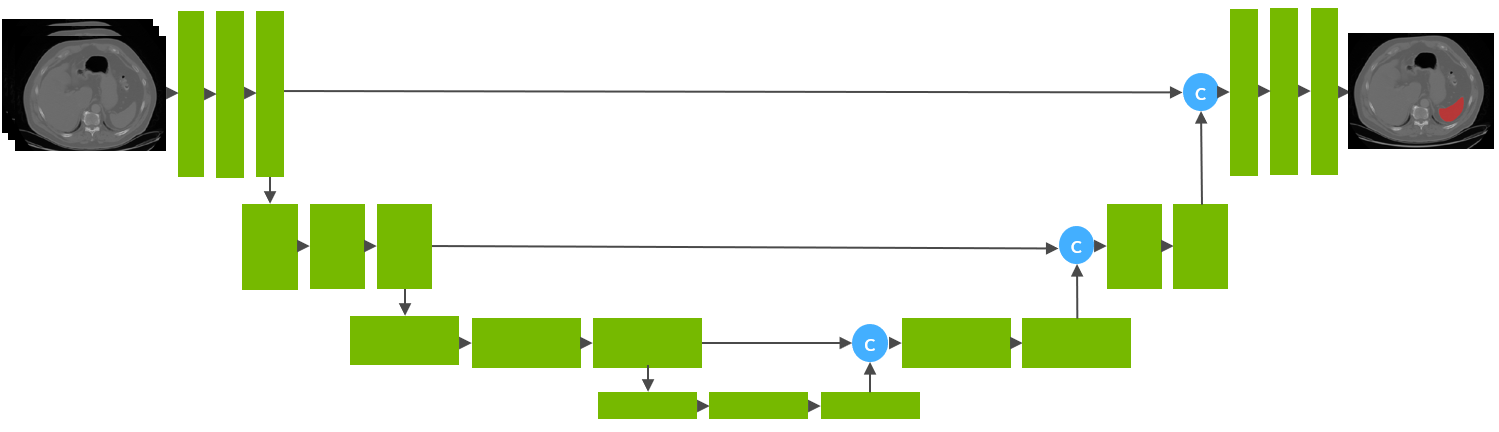
+
## Outcomes
After following along with this notebook the user will be familiar with:
diff --git a/notebooks/SpleenLiverSegmentation/SpleenSeg_Pretrained-4_27.ipynb b/notebooks/SpleenLiverSegmentation/SpleenSeg_Pretrained-4_27.ipynb
index defde63..245e2be 100644
--- a/notebooks/SpleenLiverSegmentation/SpleenSeg_Pretrained-4_27.ipynb
+++ b/notebooks/SpleenLiverSegmentation/SpleenSeg_Pretrained-4_27.ipynb
@@ -57,6 +57,16 @@
"! pip install matplotlib "
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "439cd9a3-1bb0-4ae4-b658-dbd4bf692f4b",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "! pip install --upgrade numpy"
+ ]
+ },
{
"cell_type": "code",
"execution_count": null,
@@ -403,14 +413,14 @@
"outputs": [],
"source": [
"mmar = {\n",
- " RemoteMMARKeys.ID: \"clara_pt_spleen_ct_segmentation_1\",\n",
- " RemoteMMARKeys.NAME: \"clara_pt_spleen_ct_segmentation\",\n",
+ " RemoteMMARKeys.ID: \"monai_spleen_ct_segmentation\",\n",
+ " RemoteMMARKeys.NAME: \"monai_spleen_ct_segmentation\",\n",
" RemoteMMARKeys.FILE_TYPE: \"zip\",\n",
" RemoteMMARKeys.HASH_TYPE: \"md5\",\n",
" RemoteMMARKeys.HASH_VAL: None,\n",
" RemoteMMARKeys.MODEL_FILE: os.path.join(\"models\", \"model.pt\"),\n",
- " RemoteMMARKeys.CONFIG_FILE: os.path.join(\"config\", \"config_train.json\"),\n",
- " RemoteMMARKeys.VERSION: 2,\n",
+ " RemoteMMARKeys.CONFIG_FILE: os.path.join(\"configs\", \"train.json\"),\n",
+ " RemoteMMARKeys.VERSION: \"0.5.3\"\n",
"}"
]
},
@@ -424,6 +434,17 @@
"mmar['name']"
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "5e96f360-276c-449c-b60d-fdcb1b280c92",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "from monai.apps import download_mmar\n",
+ "download_mmar(mmar['name'], mmar_dir=root_dir, version=mmar['version'])\n"
+ ]
+ },
{
"cell_type": "code",
"execution_count": null,
@@ -432,26 +453,8 @@
"outputs": [],
"source": [
"device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\") #torch.device(\"cpu\")\n",
- "if PRETRAINED:\n",
- " print(\"using a pretrained model.\")\n",
- " try: #MONAI=0.8\n",
- " unet_model = load_from_mmar(\n",
- " item = mmar['name'], \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " version=mmar['version'],\n",
- " pretrained=True)\n",
- " except: #MONAI<0.8\n",
- " unet_model = load_from_mmar(\n",
- " mmar, \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " pretrained=True)\n",
- " model = unet_model\n",
- "else: \n",
- " print(\"using a randomly init. model.\")\n",
- " model = UNet(\n",
- " dimensions=3,\n",
+ "model = UNet(\n",
+ " spatial_dims=3,\n",
" in_channels=1,\n",
" out_channels=2,\n",
" channels=(16, 32, 64, 128, 256),\n",
@@ -459,7 +462,6 @@
" num_res_units=2,\n",
" norm=Norm.BATCH,\n",
" )\n",
- "\n",
"model = model.to(device)"
]
},
@@ -711,7 +713,7 @@
"metadata": {},
"outputs": [],
"source": [
- "model.load_state_dict(torch.load('monai_data/best_metric_model_pretrained.pth'))"
+ "model.load_state_dict(torch.load('monai_data/Spleen_best_metric_model_pretrained.pth'))"
]
},
{
@@ -822,144 +824,6 @@
"#### Feel free to play around in this notebook or download it and use it where a GPU is accessible"
]
},
- {
- "cell_type": "markdown",
- "id": "896388a1",
- "metadata": {},
- "source": [
- "## Additional Exercise: Use liver segmentation in addition to spleen\n",
- " - Just need to load liver segmentation from NVIDIA\n",
- " - While we can't train this model, since we don't have training data, we can use it as a rough estimate"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "657e44a0",
- "metadata": {},
- "outputs": [],
- "source": [
- "mmarliver = {\n",
- " RemoteMMARKeys.ID: \"clara_pt_liver_and_tumor_ct_segmentation_1\",\n",
- " RemoteMMARKeys.NAME: \"clara_pt_liver_and_tumor_ct_segmentation\",\n",
- " RemoteMMARKeys.FILE_TYPE: \"zip\",\n",
- " RemoteMMARKeys.HASH_TYPE: \"md5\",\n",
- " RemoteMMARKeys.HASH_VAL: None,\n",
- " RemoteMMARKeys.MODEL_FILE: os.path.join(\"models\", \"model.pt\"),\n",
- " RemoteMMARKeys.CONFIG_FILE: os.path.join(\"config\", \"config_train.json\"),\n",
- " RemoteMMARKeys.VERSION: 1,\n",
- "}"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "a6fb0da7",
- "metadata": {},
- "outputs": [],
- "source": [
- " try: #MONAI=0.8\n",
- " unet_model = load_from_mmar(\n",
- " item = mmarliver['name'], \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " version=mmarliver['version'],\n",
- " pretrained=True)\n",
- " except: #MONAI<0.8\n",
- " unet_model = load_from_mmar(\n",
- " mmarliver, \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " pretrained=True)\n",
- " model = unet_model"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "55034354",
- "metadata": {},
- "outputs": [],
- "source": [
- "device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n",
- "\n",
- "print(\"using a pretrained model.\")\n",
- "try: #MONAI=0.8\n",
- " unet_model = load_from_mmar(\n",
- " item = mmarliver['name'], \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " version=mmarliver['version'],\n",
- " pretrained=True)\n",
- "except: #MONAI<0.8\n",
- " unet_model = load_from_mmar(\n",
- " mmarliver, \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " pretrained=True)\n",
- "model = unet_model.to(device)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "a79c1731",
- "metadata": {},
- "outputs": [],
- "source": [
- "num_classesP=3\n",
- "num_classesL=2\n",
- "post_pred = Compose([EnsureType(), AsDiscrete(argmax=True, to_onehot=num_classesP)])\n",
- "post_label = Compose([EnsureType(), AsDiscrete(to_onehot=num_classesL)])\n",
- "model.eval()\n",
- "with torch.no_grad():\n",
- " for data in DataLoader(test_ds, batch_size=1, num_workers=2):\n",
- " test_inputs, test_labels = (\n",
- " data[\"image\"].to(device),\n",
- " data[\"label\"].to(device),\n",
- " )\n",
- " roi_size = (160, 160, 160)\n",
- " sw_batch_size = 4\n",
- " test_outputs = sliding_window_inference(\n",
- " test_inputs, roi_size, sw_batch_size, model, overlap=0.5)\n",
- " test_outputsliv = [post_pred(i) for i in decollate_batch(test_outputs)] # Decollate our results\n",
- " test_labelsliv = [post_label(i) for i in decollate_batch(test_labels)]"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "c0956706",
- "metadata": {},
- "outputs": [],
- "source": [
- "sliceval = 215\n",
- "maskedliv = np.ma.masked_where(test_outputsliv[0].cpu().numpy()[1][:,:,sliceval] == 0, test_outputsliv[0].cpu().numpy()[1][:,:,sliceval])\n",
- "maskedspleen = np.ma.masked_where(test_outputsSpl[0].cpu().numpy()[1][:,:,sliceval] == 0, test_outputsSpl[0].cpu().numpy()[1][:,:,sliceval])\n",
- "fig = plt.figure(frameon=False, figsize=(7,7))\n",
- "plt.title('Pretrained Calculated Liver and spleen')\n",
- "plt.imshow(np.rot90(test_ds[0]['image'][0][:,:,sliceval]), cmap='Greys_r')\n",
- "plt.imshow(np.rot90(maskedliv), cmap='cividis', alpha=0.75)\n",
- "plt.imshow(np.rot90(maskedspleen), cmap='viridis', alpha=0.75)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "5bdfdbe9",
- "metadata": {},
- "outputs": [],
- "source": [
- "sliceval = 110\n",
- "maskedliv = np.ma.masked_where(test_outputsliv[0].cpu().numpy()[1][:,sliceval,:] == 0, test_outputsliv[0].cpu().numpy()[1][:,sliceval,:])\n",
- "maskedspleen = np.ma.masked_where(test_outputsSpl[0].cpu().numpy()[1][:,sliceval,:] == 0, test_outputsSpl[0].cpu().numpy()[1][:,sliceval,:])\n",
- "fig = plt.figure(frameon=False, figsize=(7,7))\n",
- "plt.title('Pretrained Calculated Liver and Spleen')\n",
- "plt.imshow(np.rot90(test_ds[0]['image'][0][:,sliceval,:]), cmap='Greys_r')\n",
- "plt.imshow(np.rot90(maskedliv), cmap='cividis', alpha=0.75)\n",
- "plt.imshow(np.rot90(maskedspleen), cmap='viridis', alpha=0.75)"
- ]
- },
{
"cell_type": "markdown",
"id": "bf745751",
diff --git a/notebooks/elasticBLAST/run_elastic_blast.ipynb b/notebooks/elasticBLAST/run_elastic_blast.ipynb
index ed083aa..9ff36cf 100644
--- a/notebooks/elasticBLAST/run_elastic_blast.ipynb
+++ b/notebooks/elasticBLAST/run_elastic_blast.ipynb
@@ -31,7 +31,8 @@
"metadata": {},
"source": [
"## Prerequisites\n",
- "If at any point, you get an API has not been enabled error, go to [this page](https://cloud.google.com/endpoints/docs/openapi/enable-api#console), click `Go to APIs and Services`, then search for you API and click `Enable`."
+ "* If at any point, you get an API has not been enabled error, go to [this page](https://cloud.google.com/endpoints/docs/openapi/enable-api#console), click `Go to APIs and Services`, then search for you API and click `Enable`.\n",
+ "* If you see an error indicating that the dependency 'gke-gcloud-auth-plugin' is missing, you may install the plugin using the following command `! sudo apt-get install google-cloud-cli-gke-gcloud-auth-plugin`."
]
},
{
@@ -156,6 +157,7 @@
"gcp-project = YOUR_GCP_PROJECT_ID\n",
"gcp-region = us-east4\n",
"gcp-zone = us-east4-c\n",
+ "gke-version = 1.30\n",
"\n",
"[cluster]\n",
"num-nodes = 6\n",
\n",
- " \n"
+ " "
]
},
{
diff --git a/notebooks/GenAI/example_scripts/example_langchain_chat_llama_2_zeroshot.py b/notebooks/GenAI/example_scripts/example_langchain_chat_llama_2_zeroshot.py
index e554caf..27c86cf 100644
--- a/notebooks/GenAI/example_scripts/example_langchain_chat_llama_2_zeroshot.py
+++ b/notebooks/GenAI/example_scripts/example_langchain_chat_llama_2_zeroshot.py
@@ -3,6 +3,7 @@
from langchain.prompts import PromptTemplate
#from langchain.llms import VertexAIModelGarden
from langchain_google_vertexai import ChatVertexAI
+from langchain_google_vertexai import VertexAI
import sys
import json
import os
@@ -30,7 +31,7 @@ def build_chain():
#llm = VertexAIModelGarden(project=PROJECT_ID, endpoint_id=ENDPOINT_ID, location=LOCATION_ID)
llm = VertexAI(
- model_name="chat-bison@002",
+ model_name="gemini-2.0-flash",
max_output_tokens=1024,
temperature=0.2,
top_p=0.8,
diff --git a/notebooks/GenAI/example_scripts/example_vectorsearch_chat_llama_2_zeroshot.py b/notebooks/GenAI/example_scripts/example_vectorsearch_chat_llama_2_zeroshot.py
index 85a9033..fef58ca 100644
--- a/notebooks/GenAI/example_scripts/example_vectorsearch_chat_llama_2_zeroshot.py
+++ b/notebooks/GenAI/example_scripts/example_vectorsearch_chat_llama_2_zeroshot.py
@@ -4,6 +4,7 @@
from langchain_google_vertexai import VertexAIEmbeddings
from langchain_google_vertexai import VectorSearchVectorStore
from langchain_google_vertexai import ChatVertexAI
+from langchain_google_vertexai import VertexAI
import sys
import json
import os
@@ -35,7 +36,7 @@ def build_chain():
#llm = VertexAIModelGarden(project=PROJECT_ID, endpoint_id=ENDPOINT_ID, location=LOCATION_ID)
llm = VertexAI(
- model_name="chat-bison@002",
+ model_name="gemini-2.0-flash",
max_output_tokens=1024,
temperature=0.2,
top_p=0.8,
diff --git a/notebooks/GenAI/langchain_on_vertex.ipynb b/notebooks/GenAI/langchain_on_vertex.ipynb
index 105e3ec..5dcee40 100644
--- a/notebooks/GenAI/langchain_on_vertex.ipynb
+++ b/notebooks/GenAI/langchain_on_vertex.ipynb
@@ -89,7 +89,9 @@
"from langchain_community.document_loaders import WebBaseLoader\n",
"from langchain.chains.summarize import load_summarize_chain\n",
"from langchain.schema.prompt_template import format_document\n",
- "from langchain_text_splitters import RecursiveCharacterTextSplitter"
+ "from langchain_text_splitters import RecursiveCharacterTextSplitter\n",
+ "\n",
+ "! export USER_AGENT=\"my-langchain-script/1.0\""
]
},
{
@@ -108,7 +110,7 @@
"outputs": [],
"source": [
"loader = WebBaseLoader(\"https://pubmed.ncbi.nlm.nih.gov/37883540/\")\n",
- "docs = loader.load()"
+ "docs = loader.load()\n"
]
},
{
@@ -118,7 +120,7 @@
"metadata": {},
"outputs": [],
"source": [
- "llm = ChatVertexAI()\n",
+ "llm = ChatVertexAI(model_name=\"gemini-1.5-flash-002\")\n",
"print('the LLM and default params are : ', llm)\n",
"\n",
"chain = load_summarize_chain(llm, chain_type=\"stuff\")\n",
@@ -274,7 +276,7 @@
"outputs": [],
"source": [
"# index the document using FAISS\n",
- "embeddings = VertexAIEmbeddings(model_name=\"textembedding-gecko@003\")\n",
+ "embeddings = VertexAIEmbeddings(model_name=\"text-embedding-004\")\n",
"faiss_index = FAISS.from_documents(pages, embeddings)"
]
},
@@ -331,7 +333,7 @@
" )\n",
" }\n",
" | PromptTemplate.from_template(\"Summarize the following content in around 200 words:\\n\\n{content}\")\n",
- " | ChatVertexAI()\n",
+ " | ChatVertexAI(model=\"gemini-1.5-flash-002\")\n",
" | StrOutputParser()\n",
")"
]
@@ -388,7 +390,7 @@
" )\n",
" }\n",
" | PromptTemplate.from_template(prompt_str) \n",
- " | ChatVertexAI()\n",
+ " | ChatVertexAI(model=\"gemini-1.5-flash-002\")\n",
" | StrOutputParser()\n",
")"
]
@@ -483,7 +485,7 @@
"metadata": {},
"outputs": [],
"source": [
- "llm = ChatVertexAI(model_name=\"codechat-bison@002\", max_output_tokens=1000, temperature=0.3)"
+ "llm = ChatVertexAI(model_name=\"gemini-2.0-flash-001\", max_output_tokens=1000, temperature=0.3)"
]
},
{
diff --git a/notebooks/GoogleBatch/nextflow/Part1_GBatch_Nextflow.ipynb b/notebooks/GoogleBatch/nextflow/Part1_GBatch_Nextflow.ipynb
index 9b61d1c..717365d 100644
--- a/notebooks/GoogleBatch/nextflow/Part1_GBatch_Nextflow.ipynb
+++ b/notebooks/GoogleBatch/nextflow/Part1_GBatch_Nextflow.ipynb
@@ -187,7 +187,7 @@
" ]\n",
" }\n",
" }\n",
- " ]\n",
+ " ],\n",
" \"volumes\": [\n",
" {\n",
" \"gcs\": {\n",
diff --git a/notebooks/GoogleBatch/nextflow/Part2_GBatch_Nextflow.ipynb b/notebooks/GoogleBatch/nextflow/Part2_GBatch_Nextflow.ipynb
index 1e0e8fe..debf12d 100644
--- a/notebooks/GoogleBatch/nextflow/Part2_GBatch_Nextflow.ipynb
+++ b/notebooks/GoogleBatch/nextflow/Part2_GBatch_Nextflow.ipynb
@@ -180,9 +180,26 @@
"outputs": [],
"source": [
"#Install SRAtools to download data\n",
+ "\n",
+ "#Option 1: Using mamba\n",
"! mamba install -c bioconda -c conda-forge sra-tools==2.11.0 -y"
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "1242056b-39f3-435d-b0d4-2757c3faa0ff",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#Option 2: Manual Install with wget \n",
+ "\n",
+ "!wget https://ftp-trace.ncbi.nlm.nih.gov/sra/sdk/current/sratoolkit.current-ubuntu64.tar.gz && \\\n",
+ "tar -xvzf sratoolkit.current-ubuntu64.tar.gz && \\\n",
+ "export PATH=$PATH:$(pwd)/sratoolkit.*-ubuntu64/bin && \\\n",
+ "fasterq-dump --version"
+ ]
+ },
{
"cell_type": "markdown",
"id": "1bc477a9-7b3f-431e-93b7-50e96809bfc5",
@@ -238,7 +255,7 @@
"outputs": [],
"source": [
"%%time\n",
- "! fasterq-dump -f -e 8 -m 24G SRR067701.sra"
+ "! fasterq-dump -f -e 8 -m 24G SRR067701"
]
},
{
diff --git a/notebooks/SRADownload/SRA-Download.ipynb b/notebooks/SRADownload/SRA-Download.ipynb
index 665d19b..30171a8 100644
--- a/notebooks/SRADownload/SRA-Download.ipynb
+++ b/notebooks/SRADownload/SRA-Download.ipynb
@@ -59,9 +59,17 @@
"id": "01213dae",
"metadata": {},
"source": [
- "Install dependencies, using mamba (you could also use conda). At the time of writing, the version of SRA tools available with the Anaconda distribution was v.2.11.0. If you want to install the latest version, download and install from [here](https://github.com/ncbi/sra-tools/wiki/01.-Downloading-SRA-Toolkit). If you do the direct install, you will also need to configure interactively following [this guide](https://github.com/ncbi/sra-tools/wiki/05.-Toolkit-Configuration), you can do that by opening a terminal and running the commands there."
+ "Install dependencies, using mamba (you could also use conda) or manually by downloading and installing the package. At the time of writing, the version of SRA tools available with the Anaconda distribution was v.2.11.0. If you want to install the latest version, download and install from [here](https://github.com/ncbi/sra-tools/wiki/01.-Downloading-SRA-Toolkit). If you do the direct install, you will also need to configure interactively following [this guide](https://github.com/ncbi/sra-tools/wiki/05.-Toolkit-Configuration), you can do that by opening a terminal and running the commands there."
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "3e0bb6f1-05ee-4215-94e3-8365f7550688",
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ },
{
"cell_type": "code",
"execution_count": null,
@@ -90,6 +98,19 @@
"! fasterq-dump -h"
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "5df3de21-bb3d-4d38-bcb1-30f5b10c9d01",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "!wget https://ftp-trace.ncbi.nlm.nih.gov/sra/sdk/current/sratoolkit.current-ubuntu64.tar.gz && \\\n",
+ "tar -xvzf sratoolkit.current-ubuntu64.tar.gz && \\\n",
+ "export PATH=$PATH:$(pwd)/sratoolkit.*-ubuntu64/bin && \\\n",
+ "fasterq-dump --version"
+ ]
+ },
{
"cell_type": "markdown",
"id": "7d8d3512-5307-42f7-9405-495fe1ca5be2",
diff --git a/notebooks/SpleenLiverSegmentation/README.md b/notebooks/SpleenLiverSegmentation/README.md
index 976552f..e480f8b 100644
--- a/notebooks/SpleenLiverSegmentation/README.md
+++ b/notebooks/SpleenLiverSegmentation/README.md
@@ -9,8 +9,7 @@ The Spleen model is additionally retrained on the medical decathlon spleen datas
Data is not necessary to be downloaded to run the notebook. The notebook downloads the data during it's run.
The notebook uses the Python package [MONAI](https://monai.io/), the Medical Open Network for Artificial Intelligence.
-- Spleen Model - [clara_pt_spleen_ct_segmentation_V2](https://catalog.ngc.nvidia.com/orgs/nvidia/teams/monaitoolkit/models/monai_spleen_ct_segmentation)
-- Liver Model - [clara_pt_liver_and_tumor_ct_segmentation_V1]()
+
## Outcomes
After following along with this notebook the user will be familiar with:
diff --git a/notebooks/SpleenLiverSegmentation/SpleenSeg_Pretrained-4_27.ipynb b/notebooks/SpleenLiverSegmentation/SpleenSeg_Pretrained-4_27.ipynb
index defde63..245e2be 100644
--- a/notebooks/SpleenLiverSegmentation/SpleenSeg_Pretrained-4_27.ipynb
+++ b/notebooks/SpleenLiverSegmentation/SpleenSeg_Pretrained-4_27.ipynb
@@ -57,6 +57,16 @@
"! pip install matplotlib "
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "439cd9a3-1bb0-4ae4-b658-dbd4bf692f4b",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "! pip install --upgrade numpy"
+ ]
+ },
{
"cell_type": "code",
"execution_count": null,
@@ -403,14 +413,14 @@
"outputs": [],
"source": [
"mmar = {\n",
- " RemoteMMARKeys.ID: \"clara_pt_spleen_ct_segmentation_1\",\n",
- " RemoteMMARKeys.NAME: \"clara_pt_spleen_ct_segmentation\",\n",
+ " RemoteMMARKeys.ID: \"monai_spleen_ct_segmentation\",\n",
+ " RemoteMMARKeys.NAME: \"monai_spleen_ct_segmentation\",\n",
" RemoteMMARKeys.FILE_TYPE: \"zip\",\n",
" RemoteMMARKeys.HASH_TYPE: \"md5\",\n",
" RemoteMMARKeys.HASH_VAL: None,\n",
" RemoteMMARKeys.MODEL_FILE: os.path.join(\"models\", \"model.pt\"),\n",
- " RemoteMMARKeys.CONFIG_FILE: os.path.join(\"config\", \"config_train.json\"),\n",
- " RemoteMMARKeys.VERSION: 2,\n",
+ " RemoteMMARKeys.CONFIG_FILE: os.path.join(\"configs\", \"train.json\"),\n",
+ " RemoteMMARKeys.VERSION: \"0.5.3\"\n",
"}"
]
},
@@ -424,6 +434,17 @@
"mmar['name']"
]
},
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "5e96f360-276c-449c-b60d-fdcb1b280c92",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "from monai.apps import download_mmar\n",
+ "download_mmar(mmar['name'], mmar_dir=root_dir, version=mmar['version'])\n"
+ ]
+ },
{
"cell_type": "code",
"execution_count": null,
@@ -432,26 +453,8 @@
"outputs": [],
"source": [
"device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\") #torch.device(\"cpu\")\n",
- "if PRETRAINED:\n",
- " print(\"using a pretrained model.\")\n",
- " try: #MONAI=0.8\n",
- " unet_model = load_from_mmar(\n",
- " item = mmar['name'], \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " version=mmar['version'],\n",
- " pretrained=True)\n",
- " except: #MONAI<0.8\n",
- " unet_model = load_from_mmar(\n",
- " mmar, \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " pretrained=True)\n",
- " model = unet_model\n",
- "else: \n",
- " print(\"using a randomly init. model.\")\n",
- " model = UNet(\n",
- " dimensions=3,\n",
+ "model = UNet(\n",
+ " spatial_dims=3,\n",
" in_channels=1,\n",
" out_channels=2,\n",
" channels=(16, 32, 64, 128, 256),\n",
@@ -459,7 +462,6 @@
" num_res_units=2,\n",
" norm=Norm.BATCH,\n",
" )\n",
- "\n",
"model = model.to(device)"
]
},
@@ -711,7 +713,7 @@
"metadata": {},
"outputs": [],
"source": [
- "model.load_state_dict(torch.load('monai_data/best_metric_model_pretrained.pth'))"
+ "model.load_state_dict(torch.load('monai_data/Spleen_best_metric_model_pretrained.pth'))"
]
},
{
@@ -822,144 +824,6 @@
"#### Feel free to play around in this notebook or download it and use it where a GPU is accessible"
]
},
- {
- "cell_type": "markdown",
- "id": "896388a1",
- "metadata": {},
- "source": [
- "## Additional Exercise: Use liver segmentation in addition to spleen\n",
- " - Just need to load liver segmentation from NVIDIA\n",
- " - While we can't train this model, since we don't have training data, we can use it as a rough estimate"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "657e44a0",
- "metadata": {},
- "outputs": [],
- "source": [
- "mmarliver = {\n",
- " RemoteMMARKeys.ID: \"clara_pt_liver_and_tumor_ct_segmentation_1\",\n",
- " RemoteMMARKeys.NAME: \"clara_pt_liver_and_tumor_ct_segmentation\",\n",
- " RemoteMMARKeys.FILE_TYPE: \"zip\",\n",
- " RemoteMMARKeys.HASH_TYPE: \"md5\",\n",
- " RemoteMMARKeys.HASH_VAL: None,\n",
- " RemoteMMARKeys.MODEL_FILE: os.path.join(\"models\", \"model.pt\"),\n",
- " RemoteMMARKeys.CONFIG_FILE: os.path.join(\"config\", \"config_train.json\"),\n",
- " RemoteMMARKeys.VERSION: 1,\n",
- "}"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "a6fb0da7",
- "metadata": {},
- "outputs": [],
- "source": [
- " try: #MONAI=0.8\n",
- " unet_model = load_from_mmar(\n",
- " item = mmarliver['name'], \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " version=mmarliver['version'],\n",
- " pretrained=True)\n",
- " except: #MONAI<0.8\n",
- " unet_model = load_from_mmar(\n",
- " mmarliver, \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " pretrained=True)\n",
- " model = unet_model"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "55034354",
- "metadata": {},
- "outputs": [],
- "source": [
- "device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n",
- "\n",
- "print(\"using a pretrained model.\")\n",
- "try: #MONAI=0.8\n",
- " unet_model = load_from_mmar(\n",
- " item = mmarliver['name'], \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " version=mmarliver['version'],\n",
- " pretrained=True)\n",
- "except: #MONAI<0.8\n",
- " unet_model = load_from_mmar(\n",
- " mmarliver, \n",
- " mmar_dir=root_dir,\n",
- " map_location=device,\n",
- " pretrained=True)\n",
- "model = unet_model.to(device)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "a79c1731",
- "metadata": {},
- "outputs": [],
- "source": [
- "num_classesP=3\n",
- "num_classesL=2\n",
- "post_pred = Compose([EnsureType(), AsDiscrete(argmax=True, to_onehot=num_classesP)])\n",
- "post_label = Compose([EnsureType(), AsDiscrete(to_onehot=num_classesL)])\n",
- "model.eval()\n",
- "with torch.no_grad():\n",
- " for data in DataLoader(test_ds, batch_size=1, num_workers=2):\n",
- " test_inputs, test_labels = (\n",
- " data[\"image\"].to(device),\n",
- " data[\"label\"].to(device),\n",
- " )\n",
- " roi_size = (160, 160, 160)\n",
- " sw_batch_size = 4\n",
- " test_outputs = sliding_window_inference(\n",
- " test_inputs, roi_size, sw_batch_size, model, overlap=0.5)\n",
- " test_outputsliv = [post_pred(i) for i in decollate_batch(test_outputs)] # Decollate our results\n",
- " test_labelsliv = [post_label(i) for i in decollate_batch(test_labels)]"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "c0956706",
- "metadata": {},
- "outputs": [],
- "source": [
- "sliceval = 215\n",
- "maskedliv = np.ma.masked_where(test_outputsliv[0].cpu().numpy()[1][:,:,sliceval] == 0, test_outputsliv[0].cpu().numpy()[1][:,:,sliceval])\n",
- "maskedspleen = np.ma.masked_where(test_outputsSpl[0].cpu().numpy()[1][:,:,sliceval] == 0, test_outputsSpl[0].cpu().numpy()[1][:,:,sliceval])\n",
- "fig = plt.figure(frameon=False, figsize=(7,7))\n",
- "plt.title('Pretrained Calculated Liver and spleen')\n",
- "plt.imshow(np.rot90(test_ds[0]['image'][0][:,:,sliceval]), cmap='Greys_r')\n",
- "plt.imshow(np.rot90(maskedliv), cmap='cividis', alpha=0.75)\n",
- "plt.imshow(np.rot90(maskedspleen), cmap='viridis', alpha=0.75)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "5bdfdbe9",
- "metadata": {},
- "outputs": [],
- "source": [
- "sliceval = 110\n",
- "maskedliv = np.ma.masked_where(test_outputsliv[0].cpu().numpy()[1][:,sliceval,:] == 0, test_outputsliv[0].cpu().numpy()[1][:,sliceval,:])\n",
- "maskedspleen = np.ma.masked_where(test_outputsSpl[0].cpu().numpy()[1][:,sliceval,:] == 0, test_outputsSpl[0].cpu().numpy()[1][:,sliceval,:])\n",
- "fig = plt.figure(frameon=False, figsize=(7,7))\n",
- "plt.title('Pretrained Calculated Liver and Spleen')\n",
- "plt.imshow(np.rot90(test_ds[0]['image'][0][:,sliceval,:]), cmap='Greys_r')\n",
- "plt.imshow(np.rot90(maskedliv), cmap='cividis', alpha=0.75)\n",
- "plt.imshow(np.rot90(maskedspleen), cmap='viridis', alpha=0.75)"
- ]
- },
{
"cell_type": "markdown",
"id": "bf745751",
diff --git a/notebooks/elasticBLAST/run_elastic_blast.ipynb b/notebooks/elasticBLAST/run_elastic_blast.ipynb
index ed083aa..9ff36cf 100644
--- a/notebooks/elasticBLAST/run_elastic_blast.ipynb
+++ b/notebooks/elasticBLAST/run_elastic_blast.ipynb
@@ -31,7 +31,8 @@
"metadata": {},
"source": [
"## Prerequisites\n",
- "If at any point, you get an API has not been enabled error, go to [this page](https://cloud.google.com/endpoints/docs/openapi/enable-api#console), click `Go to APIs and Services`, then search for you API and click `Enable`."
+ "* If at any point, you get an API has not been enabled error, go to [this page](https://cloud.google.com/endpoints/docs/openapi/enable-api#console), click `Go to APIs and Services`, then search for you API and click `Enable`.\n",
+ "* If you see an error indicating that the dependency 'gke-gcloud-auth-plugin' is missing, you may install the plugin using the following command `! sudo apt-get install google-cloud-cli-gke-gcloud-auth-plugin`."
]
},
{
@@ -156,6 +157,7 @@
"gcp-project = YOUR_GCP_PROJECT_ID\n",
"gcp-region = us-east4\n",
"gcp-zone = us-east4-c\n",
+ "gke-version = 1.30\n",
"\n",
"[cluster]\n",
"num-nodes = 6\n",