 +
+ 
 +
+ 
-  +
+ 
diff --git a/docs/Project.toml b/docs/Project.toml new file mode 100644 index 0000000..6de6227 --- /dev/null +++ b/docs/Project.toml @@ -0,0 +1,6 @@ +[deps] +Documenter = "e30172f5-a6a5-5a46-863b-614d45cd2de4" +ParticlesMC = "98828e2c-de49-11ef-324d-d77a22de11e4" + +[compat] +Documenter = "1.8" diff --git a/docs/make.jl b/docs/make.jl new file mode 100644 index 0000000..bfcc500 --- /dev/null +++ b/docs/make.jl @@ -0,0 +1,34 @@ +using Documenter +using ParticlesMC + +readme = read(joinpath(@__DIR__, "..", "README.md"), String) +html_part = readme[1:findlast("
", readme)[end]] +html_part = replace(html_part, r" +
++ParticlesMC is a Julia package for performing atomic and molecular Monte Carlo simulations. It is designed to be efficient and user-friendly, making it suitable for both research and educational purposes. Built on top of the Arianna module, it leverages Arianna’s Monte Carlo framework. +
+ +
+  +
+
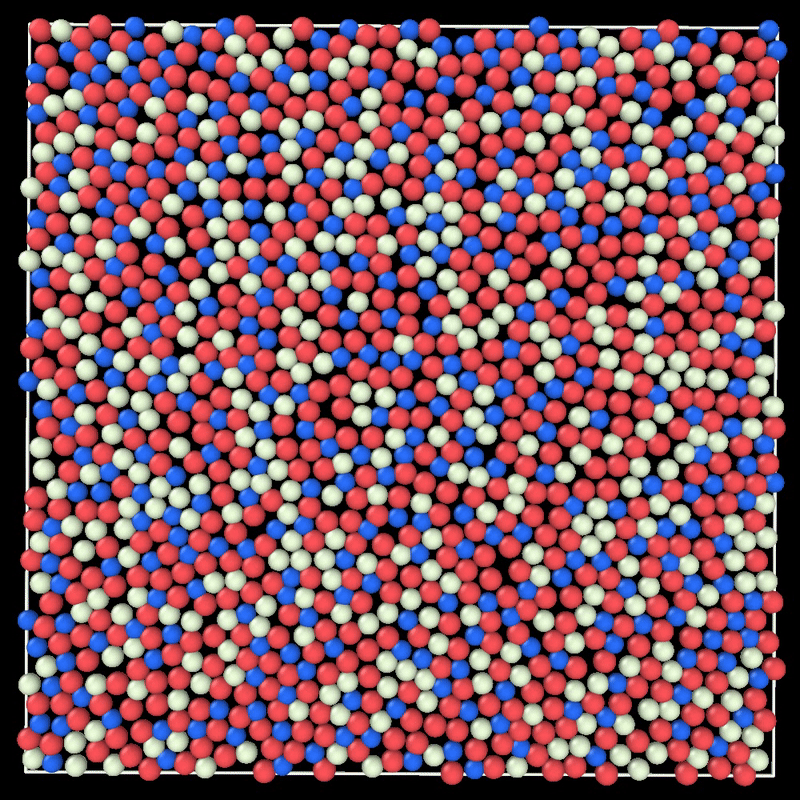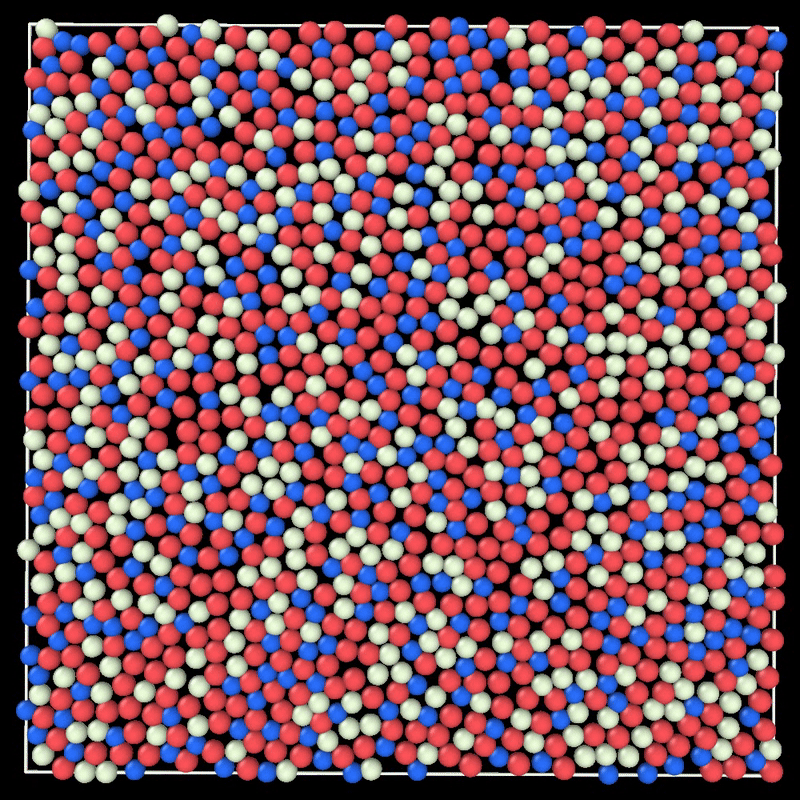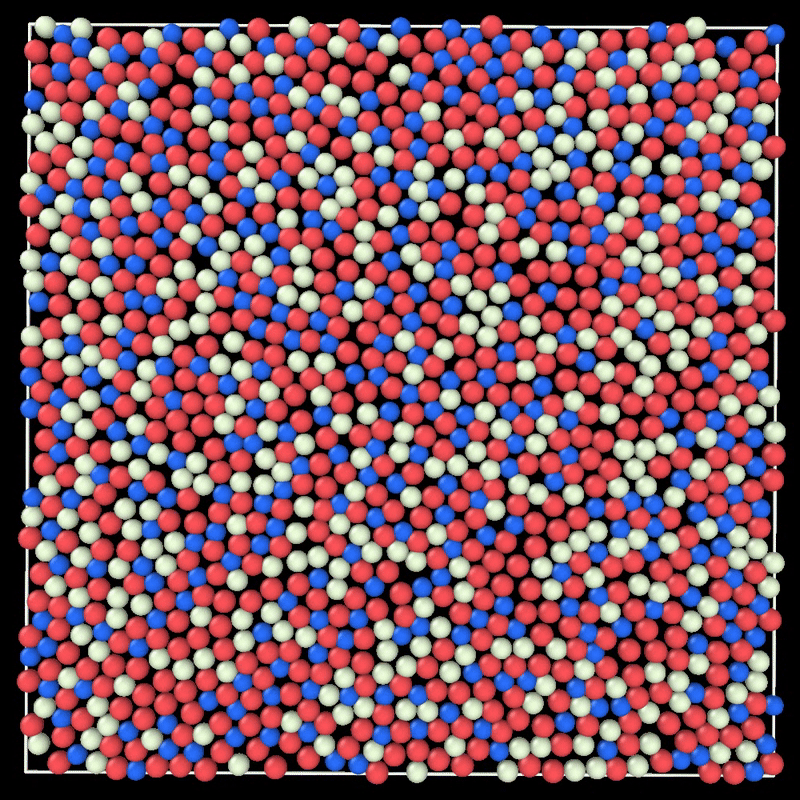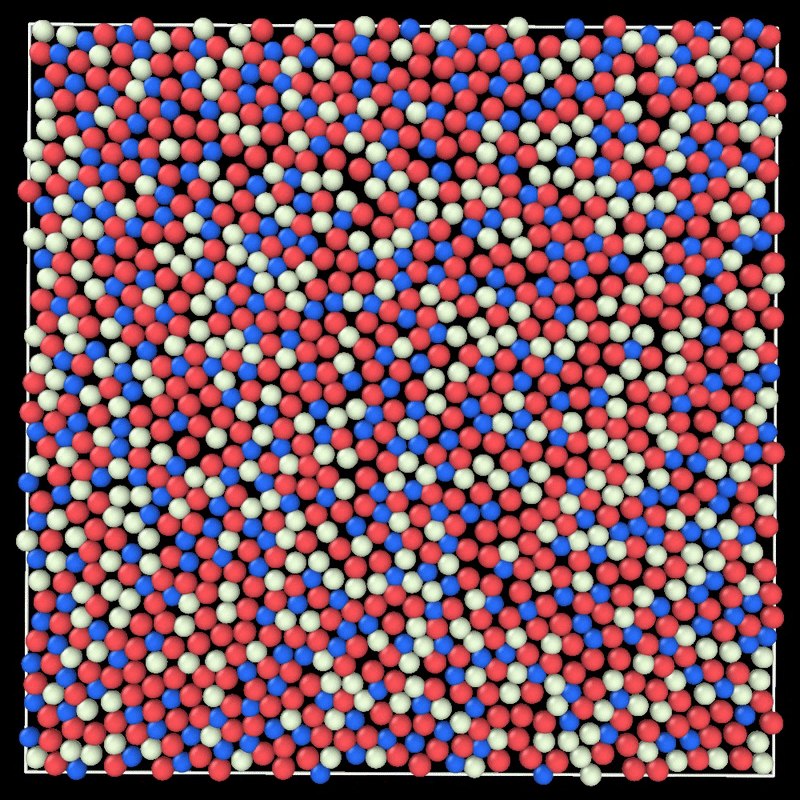
+ MC simulation of a 2D liquid. This example can be reproduced by running particlesmc params.toml in the examples/movie/ folder. Movie generated with ovito.
+
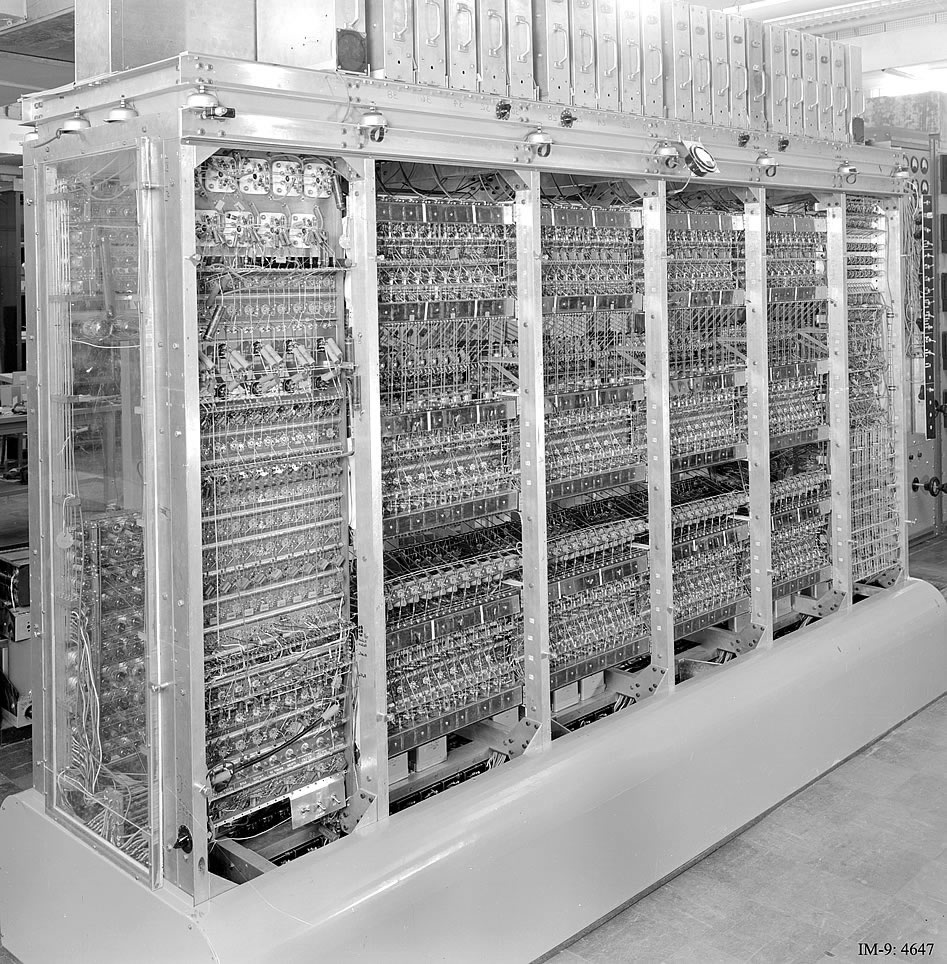 +
+The MANIAC I computer at Los Alamos National Laboratory, where Arianna Rosenbluth implemented the first MCMC algorithm. +This machine represented the cutting edge of computational capability in the 1950s. Image taken from Ref.~\cite{maniac}.
+``` + +## Monte Carlo Moves: Actions and Policies + +The transition from state $x$ to state $x'$ in the MH algorithm is commonly referred to as a *Monte Carlo move*. Monte Carlo moves can be conceptually decomposed into two components[^2]: + +- **Actions:** The type of modification applied to the system (e.g., particle displacement, rotation, swap, flip). +- **Policies:** The probabilistic rules governing how the action parameters are sampled (e.g., the distribution of displacement magnitudes and directions). Policies can be symmetric or non-symmetric. If a policy is symmetric, then the Metropolis algorithm is employed, else the Metropolis-Hastings algorithm is employed. + + +To illustrate this distinction, consider the move employed in the original Metropolis simulation. The *action* is the translation of a randomly selected particle. +To satisfy the detailed balance condition with the symmetric Metropolis acceptance rule, the translation's direction and magnitude can +be sampled from a symmetric distribution, for example uniform, Gaussian, exponential, etc. The choice of this distribution constitutes the *policy*. + +Transitions between successive states, $x_i \to x_{i+1}$, need not be governed by a single type of Monte Carlo move. In practice, it is often advantageous to define a pool of +distinct move types and to select one randomly from this pool at each step of the Markov chain. To each move type $m$ in the pool, one assigns a selection probability +$p_m$, with the normalization condition $\sum_{m} p_m = 1$. +This collection of moves, together with their associated probabilities, can be regarded as a single composite Monte Carlo move. + +## Choosing the right parameters + +```@raw html +Markov chain state space exploration for three different displacement policies. The system is a 1D particle in a potential well $U(x)$, with the target distribution being the Boltzmann distribution. The proposed move is the following: the action is the displacement of the particle and the policy is the distribution of the displacement magnitude and direction. Here, the policy is an uniform law $\sim \mathcal{U}(-u,u)$ with $u>0$. Each dot represents a state in the Markov chain, constructed from bottom to top. The horizontal position of a dot is the position of the particle. Center: When $u$ is too small, all moves are accepted but the state space is explored inefficiently due to small step sizes. Right: When $u$ is too large, nearly all moves are rejected, again leading to poor exploration. Left: An optimal choice of $u$ balances acceptance rate and step size, achieving efficient state space sampling. This illustrates the importance of tuning the proposal distribution.
+``` + +A MCMC simulation consists of two phases: a *burn-in* phase and an *equilibrium* phase[^3]. During burn-in, initial samples are discarded because the chain has not yet reached its stationary distribution. Starting from an arbitrary initial state, early samples may overweight +low-probability regions of the state space. The burn-in period must be long enough to allow the chain to converge to the target distribution before collecting samples for analysis. + +Once the chain reaches equilibrium, samples are collected to estimate properties of the system. The simulation must run long enough to +adequately explore the state space $\mathcal{X}$. However, successive samples are typically correlated, reducing the effective sample size. To assess sampling quality, one should examine autocorrelation functions and apply convergence diagnostics to verify that the chain has sufficiently explored $\mathcal{X}$[^4]. + +The choice of actions and policies is critical for efficient sampling. As shown in the above Figure, a poorly chosen policy leads to +inefficient exploration of the state space. Finding optimal actions and policies are essential for effective MCMC sampling. + +[^1]: *Equation of State Calculations by Fast Computing Machines*, Metropolis, Nicholas and Rosenbluth, Arianna W. and Rosenbluth, Marshall N. and Teller, Augusta H. and Teller, Edward, 1953 +[^2]: *Reinforcement learning: An introduction*, Sutton, Richard S and Barto, Andrew G and others, 1998 +[^3]: *Monte Carlo methods*, Guiselin, Benjamin, 2024 +[^4]: *Convergence diagnostics for markov chain monte carlo*, Roy, Vivekananda, 2020 diff --git a/docs/src/man/simulations.md b/docs/src/man/simulations.md new file mode 100644 index 0000000..a6dab42 --- /dev/null +++ b/docs/src/man/simulations.md @@ -0,0 +1,82 @@ +# Running simulations + +To perform a MC simulation of atomic or molecular systems, the procedure outlined in [Arianna](https://thedisorderedorganization.github.io/Arianna.jl/stable/#Usage) should be followed. +The first step involves initializing the system, represented by either an **Atoms** or **Molecules** structure. This requires specifying +an initial configuration (including bonding information in the molecular case), the interaction potentials between different particle types, +as well as the system’s density and temperature. +Subsequently, the desired *Arianna.jl* MCMC algorithm and the corresponding set of Monte Carlo moves are selected. +The next step consists of defining the simulation parameters, such as the total number of steps, the burn-in period, and the output directory. +Once these elements have been specified, the simulation can be executed. + +This procedure can be carried out in two ways: either through the command-line interface (CLI) or programmatically within a Julia script, +following the approach illustrated in *Arianna.jl* [examples](https://thedisorderedorganization.github.io/Arianna.jl/stable/#Usage). We detail here the CLI method, which is more user-friendly. + +## Using the command-line interface + +The CLI allows you to run simulations directly from the terminal. The basic command structure is as follows +```bash +particlesmc params.toml +``` +where **params.toml** is a parameter file written in `TOML` format that specifies the simulation settings. +The `TOML` file must provide the following information: + +- The *[system]* section specifies the input configuration file, the simulation temperature and density, and the type of neighbour list. +- The *[model]* section defines the interaction potential between particles. +- The *[simulation]* section sets the simulation type to Metropolis, the number of Monte Carlo steps, the random seed, whether to run in parallel, and the output path. +- The *[[simulation.move]]* section describes the Monte Carlo move to use. +- The *[[simulation.output]]* section configures the output (trajectories and callbacks). + +## Example: Simulating a Lennard-Jones fluid +We perform a Monte Carlo simulation of a simple Lennard-Jones fluid using the package's CLI.\@ +As explained above, we need to create a parameter file in TOML format that specifies the simulation settings. +An example of the parameter file is presented below for simulating a Lennard-Jones fluid at temperature $T=1.0$ and number density $\rho=1.0$. + +```toml +[system] +config = "config.xyz" +temperature = 1.0 +density = 1.0 +list_type = "LinkedList" + +[model] +[model."1-1"] +name = "LennardJones" +epsilon = 1.0 +sigma = 1.0 +rcut = 2.5 + +[simulation] +type = "Metropolis" +steps = 500 +output_path = "./" + +[[simulation.move]] +action = "Displacement" +probability = 1.0 +policy = "SimpleGaussian" +parameters = {sigma = 0.05} + +[[simulation.output]] +algorithm = "StoreCallbacks" +callbacks = ["energy", "acceptance"] +scheduler_params = {linear_interval = 100} + +[[simulation.output]] +algorithm = "StoreTrajectories" +scheduler_params = {linear_interval = 100} +fmt = "XYZ" +``` + +This example defines a minimal Monte Carlo simulation setup: +- The *[system]* The input configuration file is `config.xyz', the temperature is 1.0 and the density is 1.0. The neighbor list type is a linked list. +- The *[model]* The interaction potential is a Lennard-Jones potential is used for species pair `1--1` with specified parameters (`epsilon`, `sigma`, and cutoff `rcut`). +- The *[simulation]* The *Arianna.jl* MCMC method use is the Metropolis algorithm, the number of Monte Carlo steps is set to 500, and the output path is the simulation directory +- The *[[simulation.move]]* The Monte Carlo move is a displacement move with probability 1.0, guided by a simple Gaussian policy with a standard deviation (`sigma`) of 0.05. +- The *[[simulation.output]]* Two output algorithms are specified: one to store callbacks (energy and acceptance rate) every 100 steps, and another to store trajectories in XYZ format every 100 steps. + +Executing the `particlesmc params.toml` command will run a basic Metropolis Monte Carlo simulation of particles +interacting via the Lennard-Jones potential, using displacement moves, and periodically saving the system's trajectory. +When the simulation ends, a file containing a summary of the simulation will be created in the output path. + +This is a basic example of how to set up and run a Monte Carlo simulation using the package's CLI. Users can modify and extend this example to +simulate a variety of atomic and molecular systems by changing the interactions, moves, and simtulation parameters. \ No newline at end of file diff --git a/logo.png b/logo.png new file mode 100644 index 0000000..8d2b9a2 Binary files /dev/null and b/logo.png differ diff --git a/src/ParticlesMC.jl b/src/ParticlesMC.jl index c05b0ee..8120b1e 100644 --- a/src/ParticlesMC.jl +++ b/src/ParticlesMC.jl @@ -1,3 +1,9 @@ +""" +ParticlesMC: Monte Carlo simulation framework for particle systems. + +Provides core types, utilities, and the `particlesmc` command implemented with Comonicon. +Exports commonly-used types (e.g., `Particles`, `Model`) and helper functions for simulation control, I/O, and moves. +""" module ParticlesMC using Arianna, StaticArrays @@ -15,24 +21,92 @@ include("molecules.jl") include("atoms.jl") include("moves.jl") +"""Return the position of particle `i` in `system`. + +# Arguments +- `system::Particles`: the particle system +- `i::Int`: particle index + +# Returns +- Coordinates of particle `i` (e.g., an `SVector` or array). +""" get_position(system::Particles, i::Int) = @inbounds system.position[i] + +"""Return the species index (type) of particle `i`. + +# Arguments +- `system::Particles`: the particle system +- `i::Int`: particle index + +# Returns +- `Int`: species identifier of particle `i`. +""" get_species(system::Particles, i::Int) = @inbounds system.species[i] + +"""Return the interaction `Model` between the species of particles `i` and `j`. + +# Arguments +- `system::Particles`: the particle system +- `i::Int`, `j::Int`: particle indices + +# Returns +- `Model` object or callable describing pair interactions for the two species. +""" get_model(system::Particles, i::Int, j::Int) = @inbounds system.model_matrix[get_species(system, i), get_species(system, j)] + +"""Return the simulation box of `system`. + +# Returns +- Box description (usually vector or struct) representing periodic box extents. +""" get_box(system::Particles) = system.box + +"""Return the neighbour list of `system`. + +# Returns +- The neighbour list object used for pair evaluations (e.g., `NeighbourList`, `LinkedList`). +""" get_neighbour_list(system::Particles) = system.neighbour_list + +"""Return the number of particles in `system`. + +Overloads `Base.length` for `Particles`. +""" Base.length(system::Particles) = system.N + +"""Return a proper index range for `system`. + +Overloads `Base.eachindex` to allow fast indexing. +""" Base.eachindex(system::Particles) = Base.OneTo(length(system)) + +"""Return the (position, species) tuple for atom `i`. + +This overload supports indexing `atoms[i]` to get coordinates and species. +""" Base.getindex(system::Atoms, i::Int) = system.position[i], system.species[i] +"""Iterate over `Atoms` or `Molecules` returning the position and next state. + +Conforms to Julia iterator interface; yields the position of the current index. +""" function Base.iterate(system::Union{Atoms, Molecules}, state=1) state > length(system) && return nothing # Stop iteration return (system.position[state], state + 1) # Return element & next state end +"""Compute the energy contribution of particle `i` in `system` using its neighbour list. + +If a neighbour list is provided it will be used for the evaluation. +""" function compute_energy_particle(system::Particles, i::Int) return compute_energy_particle(system, i, system.neighbour_list) end +"""Compute the energy contribution for each particle index in `ids`. + +Returns an array with per-particle energies by mapping `compute_energy_particle`. +""" function compute_energy_particle(system::Particles, ids::AbstractVector) return map(i -> compute_energy_particle(system, i), ids) end @@ -40,7 +114,6 @@ end export callback_energy #export nearest_image_distance -export Model export Model, GeneralKG, JBB, BHHP, SoftSpheres, KobAndersen, Trimer export NeighbourList, LinkedList, CellList, EmptyList export Atoms, Molecules diff --git a/src/atoms.jl b/src/atoms.jl index eae23c1..b875cdc 100644 --- a/src/atoms.jl +++ b/src/atoms.jl @@ -1,3 +1,20 @@ +""" +Atoms - type representing a system of non-bonded particles. + +Description +`Atoms{D,V,C,H,T,SM} <: Particles` stores positions, species, thermodynamic +properties, and neighbour-list data needed for energy and neighbour queries. + +Fields +- `position::Vector{SVector{D,T}}`: positions of particles. +- `species::V`: species/type identifier per particle (or per-site values). +- `density::T`, `temperature::T`, `energy::Vector{T}`: thermodynamic properties. +- `model_matrix::SM`: interaction models used for pairwise energies. +- `N::Int`, `d::Int`: number of particles and dimensionality. +- `box::SVector{D,T}`: periodic box vector. +- `neighbour_list::C`: neighbour list structure (CellList/LinkedList/EmptyList). +- `species_list::H`: optional auxiliary species list structure. +""" struct Atoms{D, V<:AbstractVector, C<:NeighbourList, H, T<:AbstractFloat, SM<:AbstractArray} <: Particles position::Vector{SVector{D,T}} species::V @@ -13,6 +30,13 @@ struct Atoms{D, V<:AbstractVector, C<:NeighbourList, H, T<:AbstractFloat, SM<:Ab end +""" +Create an `Atoms` system, initialize its neighbour list, and compute initial energy. + +`System(position, species, density, temperature, model_matrix; list_type=EmptyList)` +constructs an `Atoms` instance, builds the neighbour list of type `list_type`, +and computes the initial total energy (stored in `system.energy[1]`). +""" function System(position, species, density::T, temperature::T, model_matrix; list_type=EmptyList) where{T<:AbstractFloat} @assert length(position) == length(species) N = length(position) @@ -33,6 +57,12 @@ function System(position, species, density::T, temperature::T, model_matrix; lis return system end +""" +Compute the pair energy between particles `i` and `j` for an `Atoms` system. + +Returns zero if `i == j` or if the squared distance exceeds the model's cutoff. +Otherwise returns `potential(r2, model_ij)` where `r2` is the squared nearest-image distance. +""" function compute_energy_ij(system::Atoms, i, j, position_i) # Early return using && for short-circuit evaluation i == j && return zero(typeof(system.density)) @@ -45,6 +75,12 @@ function compute_energy_ij(system::Atoms, i, j, position_i) end +""" +Compute the energy of particle `i` by brute force (no neighbour list). + +`compute_energy_particle(system, i, ::EmptyList)` sums interactions of particle `i` +with all other particles using `compute_energy_ij`. +""" function compute_energy_particle(system::Atoms, i, ::EmptyList) energy_i = zero(typeof(system.density)) position_i = get_position(system, i) @@ -55,6 +91,11 @@ function compute_energy_particle(system::Atoms, i, ::EmptyList) end +""" +Compute the energy of particle `i` using a `CellList` neighbour list. + +This restricts pair evaluations to particles in neighbouring cells of `i`. +""" function compute_energy_particle(system::Atoms, i, neighbour_list::CellList) energy_i = zero(typeof(system.density)) position_i = get_position(system, i) @@ -72,6 +113,12 @@ function compute_energy_particle(system::Atoms, i, neighbour_list::CellList) return energy_i end +""" +Compute the energy of particle `i` using a `LinkedList` neighbour list. + +This variant iterates linked list heads for neighbouring cells and accumulates +pair energies computed with `compute_energy_ij`. +""" function compute_energy_particle(system::Atoms, i, neighbour_list::LinkedList) energy_i = zero(typeof(system.density)) # Get cell of particle i diff --git a/src/molecules.jl b/src/molecules.jl index 41b1d82..b006f5f 100644 --- a/src/molecules.jl +++ b/src/molecules.jl @@ -1,3 +1,26 @@ +""" +Molecules - type managing a collection of molecules (chains) of particles. + +Description +`Molecules{D,VS,C,T,SM} <: Particles` represents a particle system organized into +molecules (or chains). It stores particle positions, molecule membership, +molecule lengths and start indices, thermodynamic properties, interaction +models, and the neighbour list structure used for energy and neighbour queries. + +Fields +- `position::Vector{SVector{D,T}}`: positions of particles. +- `species::VS`: species/type of each particle. +- `molecule::VS`: molecule identifier for each particle. +- `molecule_species::VS`: species identifier for each molecule. +- `start_mol::Vector{Int}`: starting site index of each molecule. +- `length_mol::Vector{Int}`: length (number of sites) of each molecule. +- `density::T`, `temperature::T`, `energy::Vector{T}`: thermodynamic properties. +- `model_matrix::SM`: interaction model matrix. +- `d::Int`, `N::Int`, `Nmol::Int`: dimension, number of particles, number of molecules. +- `box::SVector{D,T}`: periodic box vector. +- `neighbour_list::C`: neighbour list structure (CellList/LinkedList/EmptyList). +- `bonds::Vector{Vector{Int}}`: lists of bonded neighbours for each particle. +""" struct Molecules{D, VS<:AbstractVector, C<:NeighbourList, T<:AbstractFloat, SM<:AbstractArray} <: Particles position::Vector{SVector{D,T}} species::VS @@ -17,10 +40,39 @@ struct Molecules{D, VS<:AbstractVector, C<:NeighbourList, T<:AbstractFloat, SM< bonds::Vector{Vector{Int}} end +""" +Marker indicating that a pair of particles is bonded. +Used to select the bonded contribution (bond potentials) in energy computations. +""" struct Bonded end + +""" +Marker indicating that a pair of particles is non-bonded. + +Used to select the non-bonded pair potential contribution in energy computations. +""" struct NonBonded end +""" +Create a `Molecules` system and initialize neighbour list and energy. + +`System(position, species, molecule, density, temperature, model_matrix, bonds; molecule_species=nothing, list_type=EmptyList)` builds +and returns a `Molecules` instance. It computes `start_mol`/`length_mol`, +constructs the neighbour list of type `list_type`, builds it, and computes the +initial total energy (with a check for Inf/NaN). + +Arguments +- `position`: vector of particle positions. +- `species`: species identifier per particle. +- `molecule`: molecule id per particle. +- `density`, `temperature`: thermodynamic scalars. +- `model_matrix`: interaction models used to compute energies. +- `bonds`: bonded neighbour lists for each particle. + +Returns +- `Molecules` instance with neighbour list built and `energy[1]` set. +""" function System(position, species, molecule, density::T, temperature::T, model_matrix, bonds; molecule_species=nothing, list_type=EmptyList) where {T<:AbstractFloat} @assert length(position) == length(species) N = length(position) @@ -43,8 +95,20 @@ function System(position, species, molecule, density::T, temperature::T, model_m return system end +""" +Return the start and end indices of molecule `i` in `system`. + +`get_start_end_mol(system, i)` returns a tuple `(start, end)` for the i-th molecule, +where `start` is the first particle index and `end` is the last particle index. +""" get_start_end_mol(system::Particles, i::Int) = system.start_mol[i], system.start_mol[i] + system.length_mol[i] - 1 +""" +Return arrays of first indices and counts for consecutive blocks in `vec`. + +`get_first_and_counts(vec)` scans `vec` and returns a tuple `(firsts, counts)`, where +`firsts` are the starting indices of each run and `counts` the corresponding lengths. +""" function get_first_and_counts(vec::Vector{Int}) firsts = Int[] counts = Int[] @@ -74,6 +138,14 @@ function get_first_and_counts(vec::Vector{Int}) return firsts, counts end +""" +Check and compute the pair energy between particles `i` and `j`. + +`check_compute_energy_ij(system, i, j, position_i, bonds_i)` returns the pair energy +between `i` and `j`. If `i == j` it returns zero. It determines whether the pair +is bonded and dispatches to `compute_energy_ij` with the appropriate marker +(`Bonded` or `NonBonded`). +""" function check_compute_energy_ij(system::Molecules, i, j, position_i, bonds_i) # Early return using && for short-circuit evaluation i == j && return zero(system.density) @@ -83,7 +155,11 @@ function check_compute_energy_ij(system::Molecules, i, j, position_i, bonds_i) model_ij = get_model(system, i, j) return compute_energy_ij(system, position_i, position_j, model_ij, isbonded) end - +""" +`check_nonbonded_compute_energy_ij` returns zero if `i == j` or if `j` is bonded to `i`. +Otherwise it computes the non-bonded pair energy by dispatching to +`compute_energy_ij(..., NonBonded())`. +""" function check_nonbonded_compute_energy_ij(system::Molecules, i, j, position_i, bonds_i) # Early return using && for short-circuit evaluation i == j && return zero(system.density) @@ -108,12 +184,26 @@ function compute_energy_ij(system::Molecules, position_i, position_j, model_ij:: return bond_potential(r2, model_ij) end +""" +Compute non-bonded pair energy using pair potential with cutoff. + +This method is chosen when the pair marker is `NonBonded`. It returns zero if +the squared distance exceeds the model's cutoff squared, otherwise returns the +pair potential `potential(r2, model_ij)`. +""" function compute_energy_ij(system::Molecules, position_i, position_j, model_ij::Model, ::NonBonded) r2 = nearest_image_distance_squared(position_i, position_j, get_box(system)) cutoff2_val = cutoff2(model_ij) return r2 > cutoff2_val ? zero(typeof(system.density)) : potential(r2, model_ij) end +""" +Compute particle energy by brute force (no neighbour list). + +`compute_energy_particle(system, i, ::EmptyList)` sums interactions of particle `i` +with all particles (including bonded and non-bonded contributions via helper +functions). Used when no neighbour list is available. +""" function compute_energy_particle(system::Molecules, i, ::EmptyList) energy = zero(typeof(system.density)) position_i = system.position[i] @@ -125,6 +215,12 @@ function compute_energy_particle(system::Molecules, i, ::EmptyList) end # With linked list +""" +Compute particle energy using a `LinkedList` neighbour list. + +This variant restricts non-bonded pair evaluation to particles in neighbouring +cells defined by the linked list; bonded contributions are added explicitly. +""" function compute_energy_particle(system::Molecules, i, neighbour_list::LinkedList) energy_i = zero(typeof(system.density)) # Get cell of particle i @@ -145,6 +241,12 @@ function compute_energy_particle(system::Molecules, i, neighbour_list::LinkedLis return energy_i end +""" +Compute particle energy using a `CellList` neighbour list. + +This variant restricts non-bonded pair evaluation to particles in neighbouring +cells defined by the cell list; bonded contributions are added explicitly. +""" function compute_energy_particle(system::Molecules, i, neighbour_list::CellList) energy_i = zero(typeof(system.density)) position_i = get_position(system, i) @@ -163,6 +265,13 @@ function compute_energy_particle(system::Molecules, i, neighbour_list::CellList) return energy_i end +""" +Compute a measure of chain correlation for monodisperse chains. + +`compute_chain_correlation(system)` assumes all chains have the same length and +computes the squared sum of pairwise cross-correlations between different chain +sites over all chains (useful as an order parameter). +""" function compute_chain_correlation(system::Molecules) chain_length = system.length_mol[1] @assert all(x -> x == chain_length, system.length_mol) "All chains must have the same length" diff --git a/src/moves.jl b/src/moves.jl index 32f9af0..b357eda 100644 --- a/src/moves.jl +++ b/src/moves.jl @@ -1,5 +1,13 @@ using ComponentArrays +""" +Execute an `Action` via the Arianna API and update the system energy. + +`Arianna.perform_action!(system, action)` calls `perform_action!` to obtain the +pre- and post-action energies `(e₁, e₂)`, sets `action.δe` and updates +`system.energy[1]` accordingly (ignoring updates when energies are Inf). +It returns the tuple `(e₁, e₂)`. +""" function Arianna.perform_action!(system::Particles, action::Action) e₁, e₂ = perform_action!(system, action) if isinf(e₁) || isinf(e₂) @@ -29,10 +37,23 @@ mutable struct Displacement{T<:AbstractArray, F<:AbstractFloat} <: Action δe::F end +""" +Apply the displacement contained in `action` to the particle(s) in `system`. + +`update_position!(system, action::Displacement)` updates `system.position[action.i]` +by adding `action.δ` in-place. +""" function update_position!(system::Particles, action::Displacement) @inbounds system.position[action.i] = system.position[action.i] + action.δ end +""" +Perform a displacement action: compute pre-move energy, apply the displacement, +update the neighbour list if the particle changed cell, then compute post-move +energy and store the energy change in `action.δe`. + +Returns the tuple `(e₁, e₂)` of energies before and after the move. +""" function perform_action!(system::Particles, action::Displacement) neighbour_list = get_neighbour_list(system) e₁ = compute_energy_particle(system, action.i, neighbour_list) @@ -46,6 +67,13 @@ function perform_action!(system::Particles, action::Displacement) return e₁, e₂ end +""" +Revert a previously applied `Displacement` action. + +This function applies `action.δ` (which should typically be the inverse +displacement), restores the system energy by subtracting `action.δe`, and +updates the neighbour list if the particle changed cell. +""" function Arianna.revert_action!(system::Particles, action::Displacement) update_position!(system, action) neighbour_list = get_neighbour_list(system) @@ -56,20 +84,43 @@ function Arianna.revert_action!(system::Particles, action::Displacement) end end +""" +Invert a `Displacement` action in-place by negating its displacement vector. + +This is used to create the revert operation for a displacement move. +""" function Arianna.invert_action!(action::Displacement, ::Particles) action.δ = -action.δ end +""" +Reward function for `Displacement` under a policy-guided scheme. + +Returns the squared norm of the displacement (can be used as a positive reward +for exploration). +""" function Arianna.PolicyGuided.reward(action::Displacement, ::Particles) return norm(action.δ)^ 2 end +""" +Simple isotropic Gaussian proposal policy for displacement moves. +""" struct SimpleGaussian <: Policy end +""" +Compute the log proposal density of a `Displacement` under `SimpleGaussian`. +""" function Arianna.log_proposal_density(action::Displacement, ::SimpleGaussian, parameters, system::Particles) return -norm(action.δ)^2 / (2parameters.σ^2) - system.d * log(2π * parameters.σ^2) / 2 end +""" +Sample a `Displacement` action under `SimpleGaussian`. + +Chooses a random particle index and draws an isotropic Gaussian displacement of +dimension `system.d` with standard deviation `parameters.σ`. +""" function Arianna.sample_action!(action::Displacement, ::SimpleGaussian, parameters, system::Particles, rng) action.i = rand(rng, 1:length(system)) action.δ = rand(rng, Normal(0, parameters.σ), system.d) @@ -78,6 +129,15 @@ end ############################################################################### # DISCRETESWAP +""" +DiscreteSwap - action representing swapping species between two particles. + +Fields +- `i::Int`, `j::Int`: particle indices involved in the swap. +- `species::Tuple{Int,Int}`: pair of species identifiers being swapped. +- `particles_per_species::Tuple{Int,Int}`: population counts used for proposal densities. +- `δe::Float64`: energy difference produced by the swap. +""" mutable struct DiscreteSwap <: Action i::Int j::Int @@ -86,6 +146,14 @@ mutable struct DiscreteSwap <: Action δe::Float64 end +""" +Swap the species identifiers of particles `i` and `j` and return the total +pre- and post-swap energies for those two particles. + +`swap_particle_species!(system, spi, i, spj, j)` computes energies before the +swap, performs the species exchange, then computes energies after and returns +`(E_before, E_after)` for the pair. +""" function swap_particle_species!(system::Particles, spi, i, spj, j) e₁ᵢ = compute_energy_particle(system, i, system.neighbour_list) e₁ⱼ = compute_energy_particle(system, j, system.neighbour_list) @@ -96,12 +164,24 @@ function swap_particle_species!(system::Particles, spi, i, spj, j) return e₁ᵢ + e₁ⱼ, e₂ᵢ + e₂ⱼ end +""" +Update auxiliary species list metadata after swapping particles `i` and `j`. + +`update_species_list!(species_list, swap_species, i, j)` updates the internal +sp_ids and sp_heads structures to reflect the swap between indices `i` and `j`. +""" function update_species_list!(species_list, swap_species, i, j) species_list.sp_ids[swap_species[1]][species_list.sp_heads[i]] = j species_list.sp_ids[swap_species[2]][species_list.sp_heads[j]] = i species_list.sp_heads[i], species_list.sp_heads[j] = species_list.sp_heads[j], species_list.sp_heads[i] end +""" +Execute a `DiscreteSwap` action: swap species of particles `i` and `j`, store +`action.δe`, and update the species list auxiliary structure. + +Returns `(E_before, E_after)` for the swapped particles. +""" function perform_action!(system::Particles, action::DiscreteSwap) i, j = action.i, action.j spi, spj = get_species(system, i), get_species(system, j) @@ -111,6 +191,11 @@ function perform_action!(system::Particles, action::DiscreteSwap) return e₁, e₂ end +""" +Revert a `DiscreteSwap` action by swapping species back and restoring energy. + +This function also updates the species list metadata to undo the swap. +""" function Arianna.revert_action!(system::Particles, action::DiscreteSwap) i, j = action.i, action.j spi, spj = get_species(system, i), get_species(system, j) @@ -119,27 +204,51 @@ function Arianna.revert_action!(system::Particles, action::DiscreteSwap) system.energy[1] -= action.δe end +""" +Invert a `DiscreteSwap` action by swapping the stored particle indices. +""" function Arianna.invert_action!(action::DiscreteSwap, ::Particles) action.i, action.j = action.j, action.i end +""" +Policy-guided reward function for `DiscreteSwap` (constant reward). +""" function Arianna.PolicyGuided.reward(::DiscreteSwap, system::Particles) return one(typeof(system.temperature)) end +""" +DoubleUniform - proposal policy selecting one particle uniformly from each of two species lists. +""" struct DoubleUniform <: Policy end +""" +Log proposal density for `DiscreteSwap` under `DoubleUniform` sampling. +""" function Arianna.log_proposal_density(action::DiscreteSwap, ::DoubleUniform, parameters, system::Particles) return -log(action.particles_per_species[1] * action.particles_per_species[2]) end +""" +Sample indices for `DiscreteSwap` uniformly from the two species lists. +""" function Arianna.sample_action!(action::DiscreteSwap, ::DoubleUniform, parameters, system::Particles, rng) action.i = rand(rng, system.species_list.sp_ids[action.species[1]]) action.j = rand(rng, system.species_list.sp_ids[action.species[2]]) end +""" +EnergyBias - proposal policy biasing selection by particle energies. +""" struct EnergyBias <: Policy end +""" +Log proposal density for `DiscreteSwap` under the `EnergyBias` policy. + +This function computes the log-probability of selecting the pair `(i,j)` given +energy-weighted exponentials controlled by `parameters.θ₁` and `parameters.θ₂`. +""" function Arianna.log_proposal_density(action::DiscreteSwap, ::EnergyBias, parameters, system::Particles) energy_particle_i = compute_energy_particle(system, action.i) energy_particle_j = compute_energy_particle(system, action.j) @@ -151,6 +260,10 @@ function Arianna.log_proposal_density(action::DiscreteSwap, ::EnergyBias, parame return numerator - log_sum_exp_1 - log_sum_exp_2 end +""" +Sample a `DiscreteSwap` pair under the `EnergyBias` policy by importance sampling +based on particle energies. +""" function Arianna.sample_action!(action::DiscreteSwap, ::EnergyBias, parameters, system::Particles, rng) energy_particles_1 = compute_energy_particle(system, system.species_list.sp_ids[action.species[1]]) energy_particles_2 = compute_energy_particle(system, system.species_list.sp_ids[action.species[2]]) @@ -166,12 +279,23 @@ end ############################################################################### +""" +MoleculeFlip - action representing swapping species between two sites within the same molecule. + +Fields +- `i::Int`, `j::Int`: particle indices inside the same molecule to be swapped. +- `δe::F`: energy change due to the swap. +""" mutable struct MoleculeFlip{F<:AbstractFloat} <: Action i::Int j::Int δe::F end +""" +Perform a `MoleculeFlip` action by swapping species between two sites within a molecule. +Returns the pair energy before and after the swap `(e₁, e₂)` and stores `δe`. +""" function perform_action!(system::Particles, action::MoleculeFlip) i, j = action.i, action.j spi, spj = system.species[i], system.species[j] @@ -180,10 +304,16 @@ function perform_action!(system::Particles, action::MoleculeFlip) return e₁, e₂ end +""" +Invert a `MoleculeFlip` action by swapping stored indices. +""" function Arianna.invert_action!(action::MoleculeFlip, ::Molecules) action.i, action.j = action.j, action.i end +""" +Revert a `MoleculeFlip` action by swapping the species back and restoring energy. +""" function Arianna.revert_action!(system::Particles, action::MoleculeFlip) i, j = action.i, action.j spi, spj = get_species(system, i), get_species(system, j) @@ -191,14 +321,24 @@ function Arianna.revert_action!(system::Particles, action::MoleculeFlip) system.energy[1] -= action.δe end +""" +Reward function for `MoleculeFlip` (currently constant). +""" function reward(::MoleculeFlip, system::Particles) return one(typeof(system.temperature)) end +""" +Log proposal density for `MoleculeFlip` under `DoubleUniform` sampling (uniform between 2 choices). +""" function Arianna.log_proposal_density(action::MoleculeFlip, ::DoubleUniform, parameters, system::Particles) return -log(2) end +""" +Sample a `MoleculeFlip` action by selecting a molecule uniformly and picking two distinct +sites in that molecule with different species. +""" function Arianna.sample_action!(action::MoleculeFlip, ::DoubleUniform, parameters, system::Particles, rng) molecule_i = rand(rng, DiscreteUniform(1, system.Nmol)) start_mol, end_mol = get_start_end_mol(system, molecule_i) diff --git a/src/utils.jl b/src/utils.jl index 7b0f5df..250ba1c 100644 --- a/src/utils.jl +++ b/src/utils.jl @@ -1,6 +1,10 @@ using ConcreteStructs, Distributions, Statistics, StaticArrays +function Arianna.unnormalised_log_target_density(e, system::Particles) + return - e / system.temperature +end + function Arianna.delta_log_target_density(e1, e2, system::Particles) return -(e2 - e1) ./ system.temperature end