You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
64
64
65
-
## Sampling :earth_africa:
65
+
## Sampling 🌍
66
66
67
67
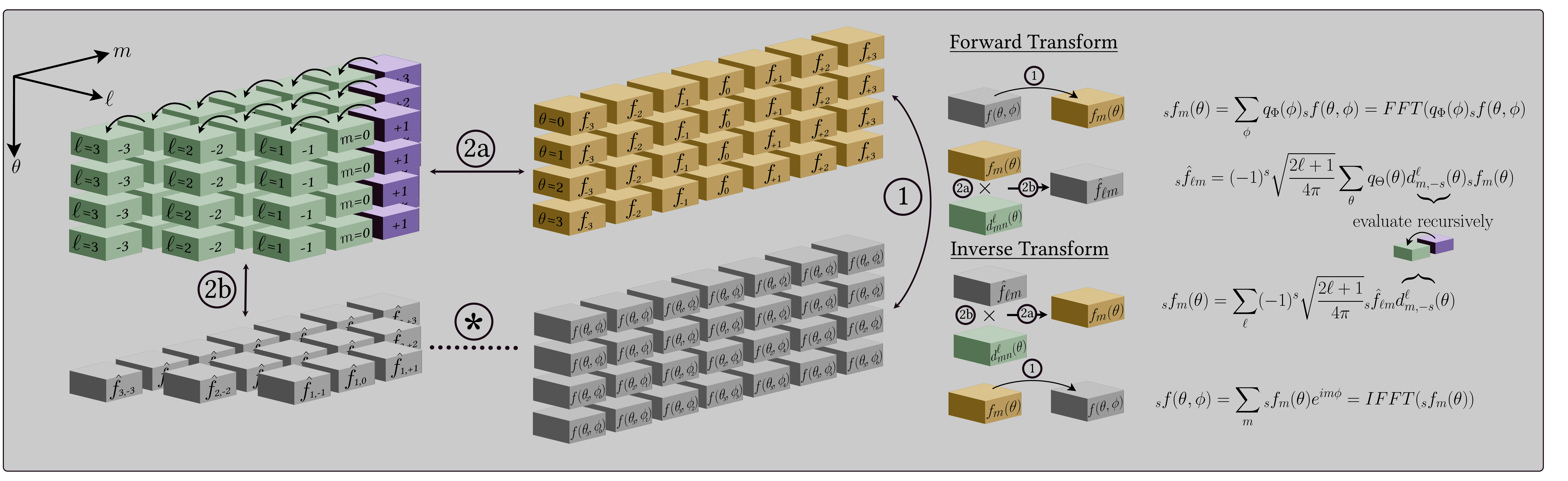
The structure of the algorithms implemented in `S2FFT` can support any
68
68
isolatitude sampling scheme. A number of sampling schemes are currently
@@ -90,7 +90,7 @@ pixels of equal areas, which has many practical advantages.
90
90
> [!NOTE]
91
91
> For algorithmic reasons JIT compilation of HEALPix transforms can become slow at high bandlimits, due to XLA unfolding of loops which currently cannot be avoided. After compiling HEALPix transforms should execute with the efficiency outlined in the associated paper, therefore this additional time overhead need only be incurred once. We are aware of this issue and are working to fix it. A fix for CPU execution has now been implemented (see example [notebook](https://astro-informatics.github.io/s2fft/tutorials/spherical_harmonic/JAX_HEALPix_backend.html)). Fix for GPU execution is coming soon.
92
92
93
-
## Installation :computer:
93
+
## Installation 💻
94
94
95
95
The Python dependencies for the `S2FFT` package are listed in the file
96
96
`requirements/requirements-core.txt` and will be automatically installed
@@ -129,7 +129,7 @@ open _build/html/index.html
129
129
> [!NOTE]
130
130
> For plotting functionality which can be found throughout our various notebooks, one must install the requirements which can be found in `requirements/requirements-plotting.txt`.
131
131
132
-
## Usage :rocket:
132
+
## Usage 🚀
133
133
134
134
To import and use `S2FFT` is as simple follows:
135
135
@@ -156,7 +156,7 @@ For further details on usage see the [documentation](https://astro-informatics.g
156
156
> [!NOTE]
157
157
> We also provide PyTorch support for the precompute version of our transforms. These are called through forward/inverse_torch(). Full PyTorch support will be provided in future releases.
158
158
159
-
## C/C++ JAX Frontends for SSHT/HEALPix :bulb:
159
+
## JAX wrappers for SSHT and HEALPix 💡
160
160
161
161
`S2FFT` also provides JAX support for existing C/C++ packages, specifically [`HEALPix`](https://healpix.jpl.nasa.gov) and [`SSHT`](https://github.com/astro-informatics/ssht). This works
162
162
by wrapping python bindings with custom JAX frontends. Note that this C/C++ to JAX interoperability is currently limited to CPU.
@@ -239,7 +239,7 @@ We encourage contributions from any interested developers. A simple
239
239
first addition could be adding support for more spherical sampling
240
240
patterns!
241
241
242
-
## Attribution :books:
242
+
## Attribution 📚
243
243
244
244
Should this code be used in any way, we kindly request that the following article is
245
245
referenced. A BibTeX entry for this reference may look like:
@@ -288,7 +288,7 @@ code builds:
288
288
}
289
289
```
290
290
291
-
## License :memo:
291
+
## License 📝
292
292
293
293
We provide this code under an MIT open-source licence with the hope that
0 commit comments