Clipping a netcdf file with Shapefile in Anaconda produces NaN values #520
Unanswered
MayMonThetKhine
asked this question in
Q&A
Replies: 1 comment
-
Perhaps a typo that you're bringing in the .shx file instead of the .shp file? |
Beta Was this translation helpful? Give feedback.
0 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
-
I want to clip my netcdf file with the shapefile, the script I run is as below. But my clipped data when I check with Panoply gives the dtr values all NaN. I attached the screenshot here.
Please enlighten me on how I can solve this one and where I went wrong. Thanks much.
==================================================================================
import xarray
import rioxarray
import geopandas
from shapely.geometry import mapping
xds = xarray.open_dataset("dtr_MON_climpact.sample_historical_NA_1970-1989.nc", drop_variables='time_bnds')
xds.rio.write_crs("EPSG:4326", inplace=True)
geodf = geopandas.read_file("Chauk_BD.shx")
clipped = xds.rio.clip(geodf.geometry.apply(mapping), geodf.crs,drop=False)
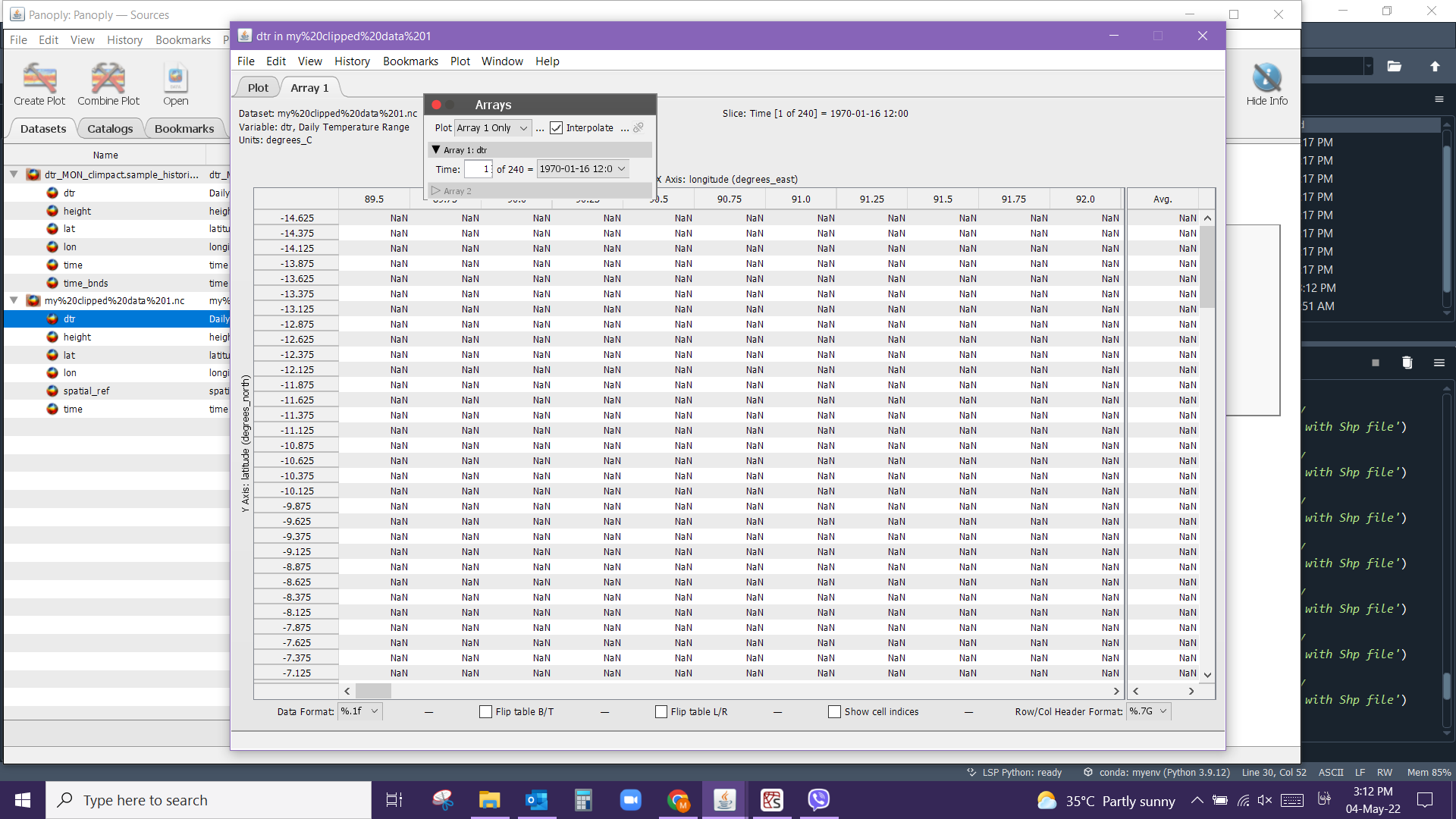
clipped.to_netcdf('my clipped data 1.nc')
Beta Was this translation helpful? Give feedback.
All reactions