Replies: 3 comments 8 replies
-
|
You may check whether your data is correct. For example, see if your forces are the gradients instead. Could you provide a correlation figure, where the x-axis represents DFT forces, and the y-axis represents forces from the model? |
Beta Was this translation helpful? Give feedback.
-
|
Hello,I've tried to train a MgCl2 solution recently. The RMSE_f_trn reached only 4e-1, when I set (CUTOFF=600, EPS_SCF=1e-5). However, when I increased CUTOFF to 800 and EPS_SCF to 1e-6, RMSE_f_trn converged to 6e-2. So whether the error of force is caused by the insufficient calculation accuracy of CP2K ? |
Beta Was this translation helpful? Give feedback.
-
|
Hi all, I am wondering if this issue is resolved. I am also seeing this same thing for my MgCl2-KCl system. Additionally, I noticed that you mentioned about multiplying cp2k forces (written in .for file) by -1. Can you help me understand why is that needed? Thanks a lot! |
Beta Was this translation helpful? Give feedback.
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
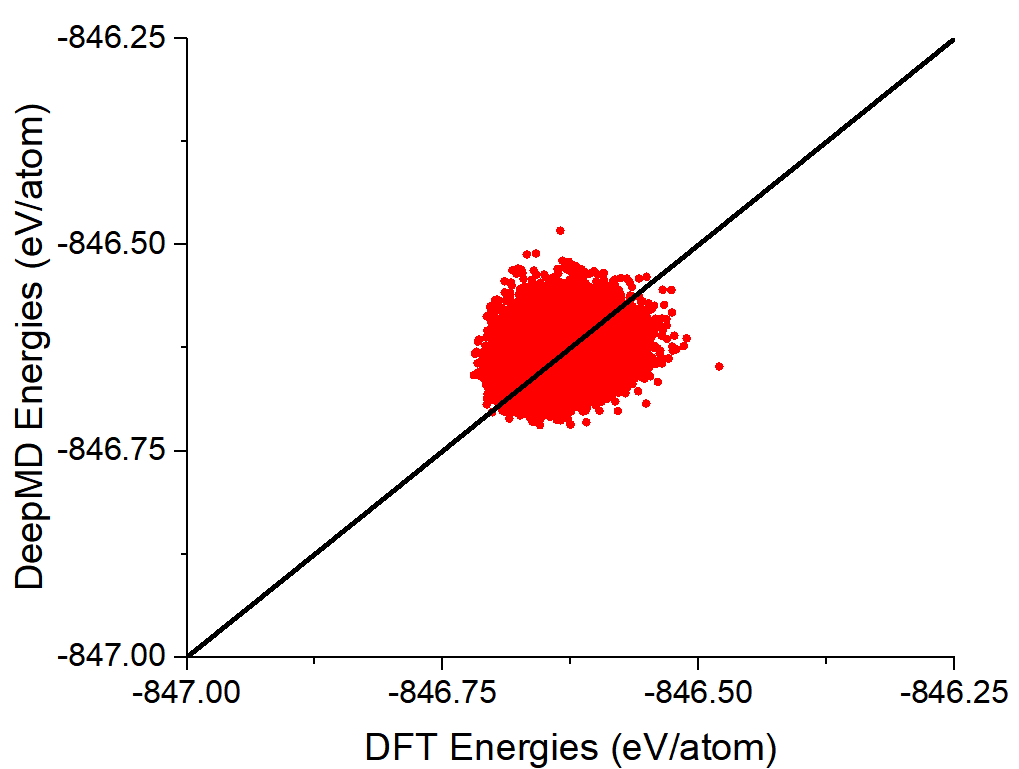
Dear all,
I plan to train the neural network for the MgCl2 (67 Mg+2 and 134 Cl-) molten salt. However, as seen below, while the RMSE for energies is reasonable, the RMSE values for forces are quite large.
For this case, training data was obtained from MD simulations at different temperatures (700 to 2000 K) and pressure conditions (1 to 100 atm), followed by DFT-derived energies and forces obtained for the snapshots using CP2K. This resulted in a total of 120000 (10% used in validation) snapshots. The "input.json" file used is as below:
{
"_comment": " model parameters",
"model": {
"type_map": ["Mg", "Cl"],
"descriptor" :{
"type": "se_e2_a",
"sel": [100, 100],
"rcut_smth": 7.80,
"rcut": 8.00,
"neuron": [25, 50, 100],
"resnet_dt": false,
"axis_neuron": 16,
"seed": 2,
"_comment": " that's all"
},
"fitting_net" : {
"neuron": [240, 240, 240],
"resnet_dt": true,
"seed": 20,
"_comment": " that's all"
},
"_comment": " that's all"
},
"learning_rate" :{
"type": "exp",
"decay_steps": 5000,
"start_lr": 0.001,
"stop_lr": 3.51e-8,
"_comment": "that's all"
},
"loss" :{
"type": "ener",
"start_pref_e": 0.02,
"limit_pref_e": 1,
"start_pref_f": 1000,
"limit_pref_f": 1,
"start_pref_v": 0,
"limit_pref_v": 0,
"_comment": " that's all"
},
"training" : {
"training_data": {
"systems": ["./mgcl2/training"],
"batch_size": "auto",
"_comment": "that's all"
},
"validation_data": {
"systems": ["./mgcl2/validation"],
"batch_size": "auto",
"numb_btch": 1,
"_comment": "that's all"
},
"numb_steps": 1000000,
"seed": 20,
"disp_file": "lcurve.out",
"disp_freq": 100,
"save_freq": 100,
"_comment": "that's all"
},
"_comment": "that's all"
}
What I have tried so far but failed to improve force RMSE values:
Surprisingly, using the same inputs and methods for the NNP for LiCl, the RMSE values are much more reasonable:
After checking the RMSE for forces more carefully, I observed that the main source of the error is from the Mg+2 ions. I further tested to see the densities obtained from 2ns trajectories (using the trained NNPs) and the densities are also very overestimated in comparison to experiments, much worse than the values obtained from AIMD (1.6007 g/cm3 for MgCl2 at 1099 K).
I was hoping if anyone could have some suggestions. Is there perhaps an error with the input files that i used?
Thank you for your time
Beta Was this translation helpful? Give feedback.
All reactions