Replies: 10 comments 5 replies
-
"sel": [432],
"rcut_smth": 5.80,
"rcut": 6.00,These settings are not good. rcut_smth is too large, to close to rcut. In addition, sel is too large. Please refer to the example setting provided with package. https://github.com/deepmodeling/deepmd-kit/blob/devel/examples/water/se_e2_a/input.json |
Beta Was this translation helpful? Give feedback.
-
|
Sorry for the late reply, some thing went wrong with my email and I have been on it. |
Beta Was this translation helpful? Give feedback.
-
|
Thank ***@***.***
How does one calculate what sel should be?
My database is simply the Silicon folder given on the deepmd website, with
9 sets, the last one for testing. It has 432 Si atoms. I counted them in
the type.raw file.
I don’t have the DFT data. I’m just beginning to learn all this, as a grad
student. I appreciate your help very much.
…On Tue, Jul 6, 2021 at 9:42 PM tuoping ***@***.***> wrote:
Sorry for the late reply, some thing went wrong with my email and I have
been on it.
"sel": [46] <---this setting is clearly not large enough. There must be
WARNINGS printed on screen or in the returned file, please take a look. A
recommended setting is "sel: [300]".
In addition, how large is your database? And what's the accuracy of your
DFT data?
—
You are receiving this because you authored the thread.
Reply to this email directly, view it on GitHub
<#807 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/ARCZHBCU6NSRJWSZFQIRUALTWOWH7ANCNFSM47OOQ7CQ>
.
|
Beta Was this translation helpful? Give feedback.
-
|
"sel" means "selected number of neighbors of center i". It doesn't mean total number of atoms in the system. "sel" is related to rcut. |
Beta Was this translation helpful? Give feedback.
-
|
Thank you @tuoping
I read that. I had calculated that for the densest silicon, there would be
45 neighbor atoms in a radius of 6A rcut. You say 46 is too small.
How did you arrive at 300 recommended for sel in this system?
I don't see any explicit way to calculate from the link you point to.
Thanks so much. 🙏
…On Thu, Jul 8, 2021 at 6:56 AM tuoping ***@***.***> wrote:
"sel" means "selected number of neighbors of center i". It doesn't mean
total number of atoms in the system. "sel" is related to rcut.
Please refer to
https://docs.deepmodeling.org/projects/deepmd/en/latest/troubleshooting/howtoset_sel.html
—
You are receiving this because you authored the thread.
Reply to this email directly, view it on GitHub
<#807 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/ARCZHBCYGDW3WSMVZTKHP3TTWV76VANCNFSM47OOQ7CQ>
.
|
Beta Was this translation helpful? Give feedback.
-
|
I apologize. Your method is definitely the best. Calculate the number of neighbors first and set "sel" according to the results. I can't see any other problem in your input file. But the error of force does look too large. I'm calling for help. @AnguseZhang |
Beta Was this translation helpful? Give feedback.
-
|
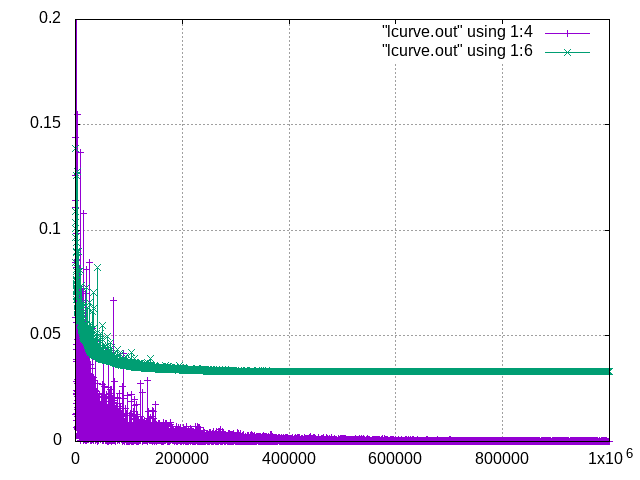
Can you use logarithmic axis for both x- and y-axis? |
Beta Was this translation helpful? Give feedback.
-
|
I don't think there is problem with training parameters. Yuzhi |
Beta Was this translation helpful? Give feedback.
-
|
May I ask which website is it? I failed to find it. |
Beta Was this translation helpful? Give feedback.
-
|
Hi! I am working on NNPs as a project. Would anyone be able to point me to a source where I can get data for Si as well as a trained NNP model? It will be really helpful. Thanks! |
Beta Was this translation helpful? Give feedback.
Uh oh!
There was an error while loading. Please reload this page.
-
Hi,
Do these look right for Si training RMS errors? Purple is energy, green is force.
Thanks
Here is the training input file:
{
"_comment": " model parameters",
"model": {
"type_map": ["Si"],
"descriptor" :{
"type": "se_a",
"sel": [432],
"rcut_smth": 5.80,
"rcut": 6.00,
"neuron": [25, 50, 100],
"resnet_dt": false,
"axis_neuron": 16,
"seed": 1,
"_comment": " that's all"
},
"fitting_net" : {
"neuron": [240, 240, 240],
"resnet_dt": true,
"seed": 1,
"_comment": " that's all"
},
"_comment": " that's all"
},
}
Beta Was this translation helpful? Give feedback.
All reactions