Converting custom NER.tsv to NER.spacy format #10905
Unanswered
shina181
asked this question in
Help: Coding & Implementations
Replies: 1 comment 6 replies
-
|
Hi @shina181, it's most likely due to your labels not being in the IOB format. How did you obtain these annotations? If it's through some software, perhaps there's an option to have it output IOB tags? Alternatively, you can rewrite the tags in place with some script (either update the TSV file itself or via |
Beta Was this translation helpful? Give feedback.
6 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
-
I have performed NER on scientific literature using some custom labels like "Operation", "target", "Amount", etc. I tried using the following command to convert my file into a corresponding json file to be further converted into a binary spacy file:
python -m spacy convert NER.tsv ./ -t json -n 1
But it is not working on my file. I have 2 columns in my file viz.tokens in the 1st column and their labels in the 2nd. Is it because my labelled tags are not in the IOB/BILOU format?
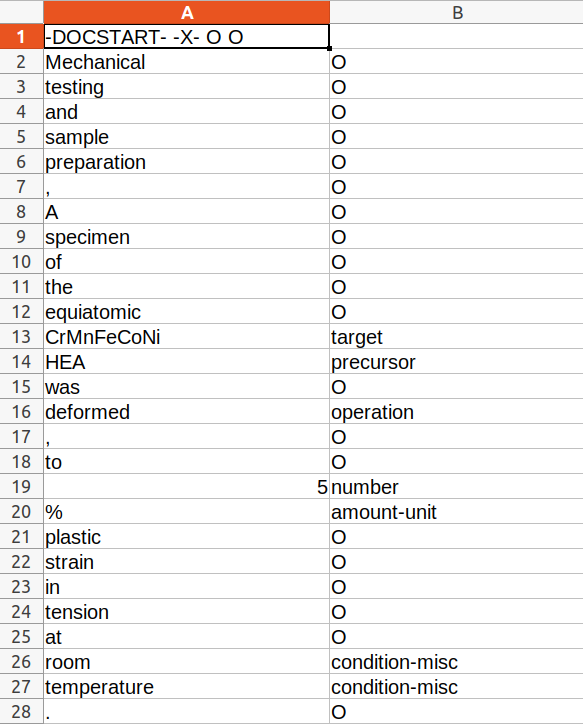
In that case how should I go about converting my file into a binary spacy file?
Should I write a code to convert the file into a json file first and then convert it into a spacy file or is there a way to directly convert my NER.tsv file to a NER.spacy file??
Thanks
Beta Was this translation helpful? Give feedback.
All reactions