-
Notifications
You must be signed in to change notification settings - Fork 5
Description
Hey @igordot , hope all is well with you!
My colleagues and I noticed something odd in the Canis lupus familiaris orthologs in orthogene, and I traced it back to babelgene. See here for the full details: neurogenomics/orthogene#30
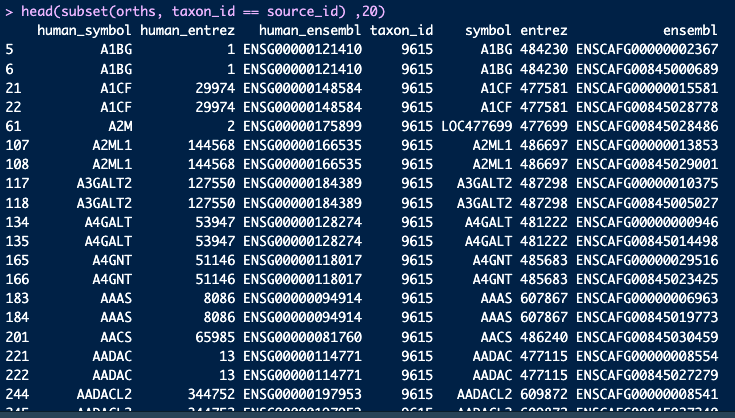
In the "symbol" column, I expect this to be the gene symbol as in the format of the origin species (in this case dog). This seems to be the case for other species like mouse, but not for dog.
source_id <- 9615
orths <- babelgene:::orthologs_df
orths <- subset(orths, taxon_id == source_id)
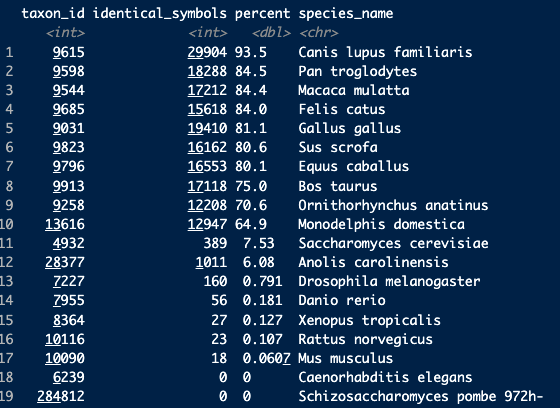
head(orths, 20)Delving a bit deeper, i checked the number of human gene symbols that are identical to their non-human orthologs in each species. This varies quite a bit across species (even within mammals), but I was wondering if this is expected? I'm familiar with the different casing with mouse genes, but not sure what the convention is for the other species.
counts <- dplyr::group_by(babelgene:::orthologs_df, taxon_id) |>
dplyr::summarise(identical_symbols=sum(human_symbol==symbol, na.rm = TRUE),
percent=sum(human_symbol==symbol, na.rm = TRUE)/dplyr::n()*100) |>
dplyr::arrange(dplyr::desc(percent))
counts$species_name <- orthogene::map_species(counts$taxon_id, method = "babelgene")
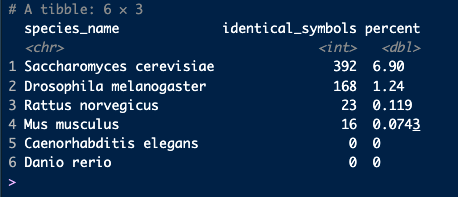
countsRepeated using the agr_orthologs_df data:
counts_agr <- dplyr::group_by(babelgene:::agr_orthologs_df, species_name) |>
dplyr::summarise(identical_symbols=sum(human_symbol==species_symbol),
percent=sum(human_symbol==species_symbol)/dplyr::n()*100) |>
dplyr::arrange(dplyr::desc(percent))
counts_agrThanks!,
Brian
Session info
Using babelgene v22.9
Details
``` R version 4.2.1 (2022-06-23) Platform: x86_64-apple-darwin17.0 (64-bit) Running under: macOS Ventura 13.2.1Matrix products: default
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggtree_3.6.2 orthogene_1.5.1
loaded via a namespace (and not attached):
[1] utf8_1.2.3 devoptera_0.99.0 R.utils_2.12.2
[4] RUnit_0.4.32 tidyselect_1.2.0 RSQLite_2.3.0
[7] AnnotationDbi_1.60.2 htmlwidgets_1.6.2 grid_4.2.1
[10] combinat_0.0-8 devtools_2.4.5 munsell_0.5.0
[13] codetools_0.2-19 DT_0.27 miniUI_0.1.1.1
[16] withr_2.5.0 colorspace_2.1-0 Biobase_2.58.0
[19] filelock_1.0.2 TreeTools_1.9.1 knitr_1.42
[22] rstudioapi_0.14 stats4_4.2.1 ggsignif_0.6.4
[25] MatrixGenerics_1.10.0 Rdpack_2.4 labeling_0.4.2
[28] GenomeInfoDbData_1.2.9 mnormt_2.1.1 optimParallel_1.0-2
[31] topGO_2.50.0 bit64_4.0.5 farver_2.1.1
[34] rprojroot_2.0.3 coda_0.19-4 vctrs_0.6.1
[37] treeio_1.23.1 generics_0.1.3 clusterGeneration_1.3.7
[40] xfun_0.37 BiocFileCache_2.6.1 R6_2.5.1
[43] doParallel_1.0.17 GenomeInfoDb_1.34.9 rsvg_2.4.0
[46] grImport2_0.2-0 bitops_1.0-7 cachem_1.0.7
[49] gridGraphics_0.5-1 DelayedArray_0.24.0 promises_1.2.0.1
[52] scales_1.2.1 gtable_0.3.3 biocViews_1.66.3
[55] processx_3.8.0 phangorn_2.11.1 ontologyPlot_1.6
[58] rlang_1.1.0 scatterplot3d_0.3-43 rstatix_0.7.2
[61] lazyeval_0.2.2 broom_1.0.4 BiocManager_1.30.20
[64] yaml_2.3.7 abind_1.4-5 ggimage_0.3.1.002
[67] backports_1.4.1 httpuv_1.6.9 RBGL_1.74.0
[70] tools_4.2.1 usethis_2.1.6 ggplotify_0.1.0
[73] ggplot2_3.4.1 ellipsis_0.3.2 paintmap_1.0
[76] RColorBrewer_1.1-3 BiocGenerics_0.44.0 sessioninfo_1.2.2
[79] Rcpp_1.0.10 plyr_1.8.8 base64enc_0.1-3
[82] zlibbioc_1.44.0 purrr_1.0.1 RCurl_1.98-1.10
[85] ps_1.7.3 prettyunits_1.1.1 ggpubr_0.6.0
[88] urlchecker_1.0.1 S4Vectors_0.36.2 SummarizedExperiment_1.28.0
[91] grr_0.9.5 fs_1.6.1 here_1.0.1
[94] magrittr_2.0.3 data.table_1.14.8 magick_2.7.4
[97] SparseM_1.81 R.cache_0.16.0 matrixStats_0.63.0
[100] pkgload_1.3.2 patchwork_1.1.2 mime_0.12
[103] evaluate_0.20 xtable_1.8-4 XML_3.99-0.14
[106] jpeg_0.1-10 IRanges_2.32.0 compiler_4.2.1
[109] tibble_3.2.1 maps_3.4.1 crayon_1.5.2
[112] R.oo_1.25.0 htmltools_0.5.4 ggfun_0.0.9
[115] later_1.3.0 tidyr_1.3.0 aplot_0.1.10
[118] expm_0.999-7 ontoProc_1.20.0 DBI_1.1.3
[121] gprofiler2_0.2.1 dbplyr_2.3.2 MASS_7.3-58.3
[124] rappdirs_0.3.3 babelgene_22.9 Matrix_1.5-3
[127] car_3.1-1 piggyback_0.1.4 cli_3.6.1
[130] quadprog_1.5-8 R.methodsS3_1.8.2 rbibutils_2.2.13
[133] parallel_4.2.1 igraph_1.4.1 GenomicRanges_1.50.2
[136] pkgconfig_2.0.3 numDeriv_2016.8-1.1 plotly_4.10.1
[139] foreach_1.5.2 stringdist_0.9.10 XVector_0.38.0
[142] BiocCheck_1.34.3 yulab.utils_0.0.6 stringr_1.5.0
[145] callr_3.7.3 digest_0.6.31 phytools_1.5-1
[148] graph_1.76.0 Biostrings_2.66.0 rmarkdown_2.20.1
[151] fastmatch_1.1-3 tidytree_0.4.2 curl_5.0.0
[154] shiny_1.7.4 lifecycle_1.0.3 nlme_3.1-162
[157] jsonlite_1.8.4 carData_3.0-5 OmaDB_2.14.0
[160] viridisLite_0.4.1 fansi_1.0.4 pillar_1.9.0
[163] ontologyIndex_2.10 lattice_0.20-45 homologene_1.4.68.19.3.27
[166] KEGGREST_1.38.0 fastmap_1.1.1 httr_1.4.5
[169] plotrix_3.8-2 pkgbuild_1.4.0 GO.db_3.16.0
[172] interactiveDisplayBase_1.36.0 glue_1.6.2 remotes_2.4.2
[175] png_0.1-8 iterators_1.0.14 BiocVersion_3.16.0
[178] bit_4.0.5 Rgraphviz_2.42.0 stringi_1.7.12
[181] profvis_0.3.7 blob_1.2.4 AnnotationHub_3.6.0
[184] rphylopic_1.0.0 memoise_2.0.1 dplyr_1.1.1
[187] ape_5.7-1
</details>