-
|
I've spent the past few months developing an astrophysics code in JAX. It works really well in forward mode and although the compilation time is a bit long (~1min) I'm very happy with the execution time. The most important bit of the code is here and the key function is The problem is that I when I try to execute this function repeatedly within The jaxpr for the gradient evaluation is ~80k lines compared to ~50k lines for the function itself and I'm using the CPU backend. Here is a notebook which reproduces the issue. I'm not sure what to do here, I've spent a lot of time optimizing each individual function and I've used I'd really appreciate any help with this! |
Beta Was this translation helpful? Give feedback.
Replies: 1 comment 13 replies
-
|
I managed to make a slightly smaller reproducible example of the problem. The issue is that the memory usage for reverse mode autodiff explodes. from functools import partial
import numpy as np
import jax.numpy as jnp
from jax import random, jit, vmap
from jax import jacfwd, jacrev
from jax import lax
from jax import make_jaxpr
from jax.config import config
config.update("jax_enable_x64", True)
config.update('jax_platform_name', 'gpu')
@jit
def lens_eq(z):
zbar = jnp.conjugate(z)
return z - 1. / zbar
@jit
def lens_eq_det_jac(z):
zbar = jnp.conjugate(z)
return 1.0 - 1.0 / jnp.abs(zbar**2)
@jit
def images_point_source(w):
w_abs_sq = jnp.abs(w) ** 2
w_bar = jnp.conjugate(w)
# Compute the image locations using the quadratic formula
z1 = (w_abs_sq + jnp.sqrt(w_abs_sq**2 + 4 * w_abs_sq)) / (2 * w_bar)
z2 = (w_abs_sq - jnp.sqrt(w_abs_sq**2 + 4 * w_abs_sq)) / (2 * w_bar)
z = jnp.stack(jnp.array([z1, z2]))
return z, jnp.ones(z.shape).astype(bool)
@jit
def mag_point_source(w):
images, mask = images_point_source(w)
det = lens_eq_det_jac(images)
mag = (1.0 / jnp.abs(det)) * mask
return mag.sum(axis=0).reshape(w.shape)
@partial(jit, static_argnames=("npts_init", "niter",))
def _images_of_source_limb(
w_center,
rho,
npts_init=500,
niter=2,
):
# Initial sampling on the source limb
theta = jnp.linspace(-np.pi, np.pi, npts_init - 1, endpoint=False)
theta = jnp.pad(theta, (0, 1), constant_values=np.pi - 1e-05)
images, mask_images = images_point_source(
rho * jnp.exp(1j * theta) + w_center,
)
det = lens_eq_det_jac(images)
parity = jnp.sign(det)
mag = jnp.sum((1.0 / jnp.abs(det)) * mask_images, axis=0)
# Refine sampling by placing geometrically fewer points each iteration
# in the regions where the magnification gradient is largest
npts_list = np.geomspace(2, npts_init, niter, endpoint=False, dtype=int)[::-1]
key = random.PRNGKey(42)
for _npts in npts_list:
# Resample theta
delta_mag = jnp.gradient(mag)
idcs_maxdelta = jnp.argsort(jnp.abs(delta_mag))[::-1][:_npts]
theta_patch = 0.5 * (theta[idcs_maxdelta] + theta[idcs_maxdelta + 1])
# Make sure that there are no exact duplicate values in `theta_patch`
# and no common values with `theta`
mask_duplicate = jnp.ones(len(theta_patch), dtype=bool)
mask_duplicate = mask_duplicate.at[
jnp.unique(theta_patch, return_index=True, size=len(theta_patch))[1]
].set(False)
mask_common = jnp.isin(theta_patch, theta, assume_unique=True)
mask = jnp.logical_or(mask_duplicate, mask_common)
theta_patch += mask * random.uniform(
key, theta_patch.shape, maxval=1e-05
) # small perturbation
images_patch, mask_images_patch = images_point_source(
rho * jnp.exp(1j * theta_patch) + w_center,
)
det_patch = lens_eq_det_jac(images_patch)
mag_patch = jnp.sum((1.0 / jnp.abs(det_patch)) * mask_images_patch, axis=0)
theta = jnp.concatenate([theta, theta_patch])
sorted_idcs = jnp.argsort(theta)
theta = theta[sorted_idcs]
mag = jnp.concatenate([mag, mag_patch])[sorted_idcs]
images = jnp.hstack([images, images_patch])[:, sorted_idcs]
mask_images = jnp.hstack([mask_images, mask_images_patch])[:, sorted_idcs]
det = jnp.hstack([det, det_patch])[:, sorted_idcs]
parity = jnp.sign(det)
return images, mask_images, parity
@jit
def _brightness_profile(z, rho, w_center, u1=0.0):
# return jnp.ones_like(z)
w = lens_eq(z)
r = jnp.abs(w - w_center) / rho
def safe_for_grad_sqrt(x):
return jnp.sqrt(jnp.where(x > 0.0, x, 0.0))
B_r = jnp.where(
r <= 1.0,
1 + safe_for_grad_sqrt(1 - r**2),
1 - safe_for_grad_sqrt(1 - 1.0 / r**2),
)
I = 3.0 / (3.0 - u1) * (u1 * B_r + 1.0 - 2.0 * u1)
return I
@jit
def simpson_quadrature(x, y):
# len(x) must be odd
h = (x[-1] - x[0]) / (len(x) - 1)
return h/3.*jnp.sum(y[0:-1:2] + 4*y[1::2] + y[2::2], axis=0)
@partial(jit, static_argnames=("npts"))
def _integrate_ld(
w_center, rho, contours, parity, tail_idcs, u1=0.0, npts=201,
):
def P(x0, y0, xl, yl):
# Construct grid in z2 and evaluate the brightness profile at each point
y = jnp.linspace(y0 * jnp.ones_like(xl), yl, npts)
integrands = _brightness_profile(
xl + 1j * y, rho, w_center, u1=u1,
)
I = simpson_quadrature(y, integrands)
return -0.5 * I
def Q(x0, y0, xl, yl):
# Construct grid in z1 and evaluate the brightness profile at each point
x = jnp.linspace(x0 * jnp.ones_like(xl), xl, npts)
integrands = _brightness_profile(
x + 1j * yl, rho, w_center, u1=u1,
)
I = simpson_quadrature(x, integrands)
return 0.5 * I
# We choose the centroid of each contour to be lower limit for the P and Q
# integrals
z0 = vmap(lambda idx, contour: contour.sum() / (idx + 1))(tail_idcs, contours)
x0, y0 = jnp.real(z0), jnp.imag(z0)
# Select k and (k + 1)th elements
contours_k = contours
contours_kp1 = jnp.pad(contours[:, 1:], ((0, 0), (0, 1)))
contours_k = vmap(lambda idx, contour: contour.at[idx].set(0.0))(
tail_idcs, contours_k
)
# Compute the integral using the midpoint rule
x_k = jnp.real(contours_k)
y_k = jnp.imag(contours_k)
x_kp1 = jnp.real(contours_kp1)
y_kp1 = jnp.imag(contours_kp1)
delta_x = x_kp1 - x_k
delta_y = y_kp1 - y_k
x_mid = 0.5 * (x_k + x_kp1)
y_mid = 0.5 * (y_k + y_kp1)
Pmid = vmap(P)(x0, y0, x_mid, y_mid)
Qmid = vmap(Q)(x0, y0, x_mid, y_mid)
I1 = Pmid * delta_x
I2 = Qmid * delta_y
mag = jnp.sum(I1 + I2, axis=1) / (np.pi * rho**2)
# sum magnifications for each image, taking into account the parity of each
# image
return jnp.abs(jnp.sum(mag * parity))
@partial(jit, static_argnames=("npts_limb", "niter_limb","npts_ld",))
def mag_extended_source(w, rho, u1=0.0, npts_limb=300, niter_limb=8, npts_ld=601):
images, images_mask, images_parity = _images_of_source_limb(
w,
rho,
npts_init=npts_limb,
niter=niter_limb,
)
# Per image parity
parity = images_parity[:, 0]
# Set last point to be equal to first point
contours = jnp.hstack([images, images[:, 0][:, None]])
tail_idcs = jnp.array([images.shape[1] - 1, images.shape[1] - 1])
return _integrate_ld(w, rho, contours, parity, tail_idcs, u1=u1, npts=npts_ld)
@jit
def _extended_source_test(w, rho):
# Magnification
u = jnp.abs(w)
mu_ps = (u ** 2 + 2) / (u * jnp.sqrt(u ** 2 + 4))
mask_valid = jnp.abs(w) > 2 * rho
return mask_valid, mu_ps
@partial(jit,static_argnames=("npts_limb","niter_limb","npts_ld",))
def mag(w_points, rho, u1=0.0, npts_limb=300, niter_limb=1, npts_ld=501):
cond, mu_approx = _extended_source_test(w_points, rho)
def body_fn(_, x):
w, c, _mu_approx = x
mag = lax.cond(
c,
lambda _: _mu_approx,
lambda w: mag_extended_source(
w,
rho,
u1=u1,
npts_limb=npts_limb,
niter_limb=niter_limb,
npts_ld=npts_ld,
),
w,
)
return 0, mag
return lax.scan(body_fn, 0, [w_points, cond, mu_approx])[1]This will crash on both the CPU and the GPU because the required memory is something like 30gb: @jit
def test_fn(params):
t0, ln_tE, u0, rho = params
t = jnp.linspace(-150, 150, 400)
tE = jnp.exp(ln_tE)
w_points = u0*1j + (t - t0) / tE
return mag(w_points, rho, u1=0., npts_limb=300, niter_limb=1).sum()
params = jnp.array([6.2, 4.39, 0.01, 0.15])
J = jit(jacrev(test_fn))
J(params)I haven't yet figured out if this is a bug in JAX or if the computational graph of my function is so large that reverse mode autodiff is supposed to be using this much memory. EDIT: see the answer by @froystig below. |
Beta Was this translation helpful? Give feedback.
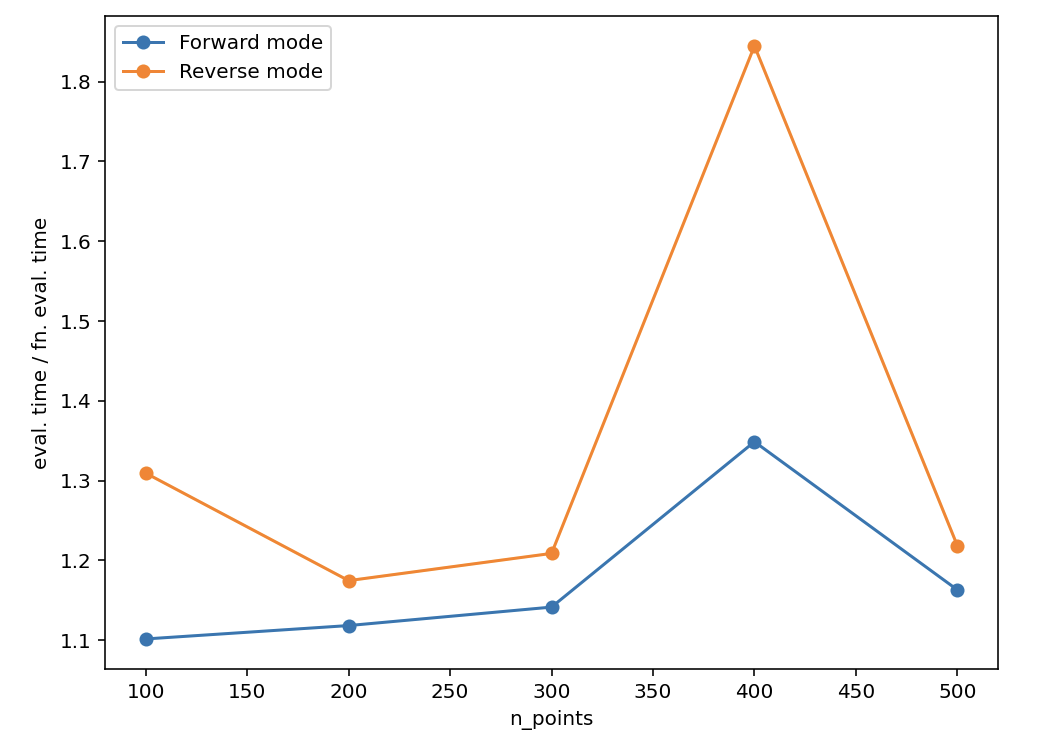
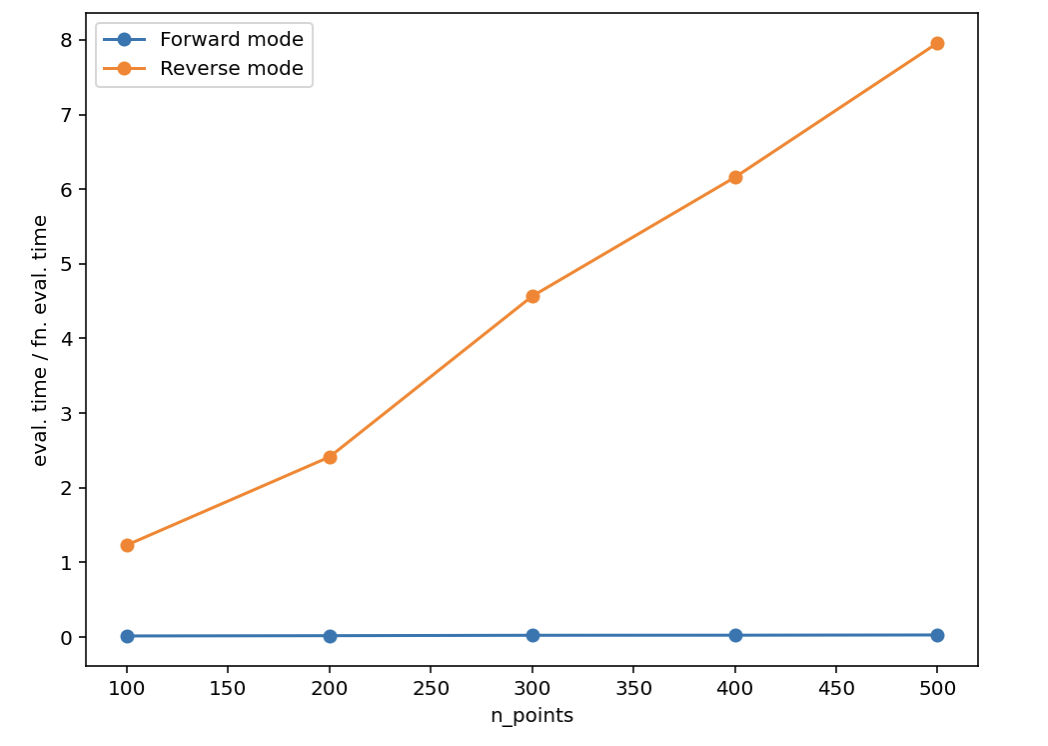
I managed to make a slightly smaller reproducible example of the problem. The issue is that the memory usage for reverse mode autodiff explodes.