-
Notifications
You must be signed in to change notification settings - Fork 5.4k
pytorch new tdnnf structure #3923
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
| self.prefinal_l = OrthonormalLinear(dim=hidden_dim, | ||
| bottleneck_dim=bottleneck_dim * 2, | ||
| time_stride=0) | ||
| bottleneck_dim=bottleneck_dim * 2) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
mm.. I'm a bit surprised this * 2 is here?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
mm.. I think you were assuming that the final layer's bottleneck is always twice the TDNN-F layers' bottleneck.
In fact we generally leave the final layer's bottleneck at 256, which for some reason seems to work across a range
of conditions. You could make that a separate configuration value.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
when I have checked the param shape of kaldi's model, I don't find the difference betwieen the final layer and previous layers what you said.
'tdnnf12.linear':'time_offsets': array([-3, 0]), 'params': (128, 2048)
'tdnnf12.affine':'time_offsets': array([0, 3]), 'params': (1024, 256)
'tdnnf13.linear':'time_offsets': array([-3, 0]), 'params': (128, 2048)
'tdnnf13.affine':'time_offsets': array([0, 3]), 'params': (1024, 256)
'prefinal-l':'params': (256, 1024)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
mm.. I'm a bit surprised this
* 2is here?
* 2 is used here to follow what kaldi does.
I've changed it to be configurable in this pullrequest: #3925
| x_left = x[:, :, :cur_context] | ||
| x_mid = x[:, :, cur_context:-cur_context:self.frame_subsampling_factor] | ||
| x_right = x[:, :, -cur_context:] | ||
| x = torch.cat([x_left, x_mid, x_right], dim=2) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm surprised that you are doing this manually rather than using a 1d convolution. This could be quite slow.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I want subsample the length of window only rather than left_context and right_context. And this is slower than before training, but it worked. please help me to write this 1d convolution.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
What might have happened here is that you tripled the dimension in the middle of the network.
This would have led to a system with many more parameters for your "dilation" system.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
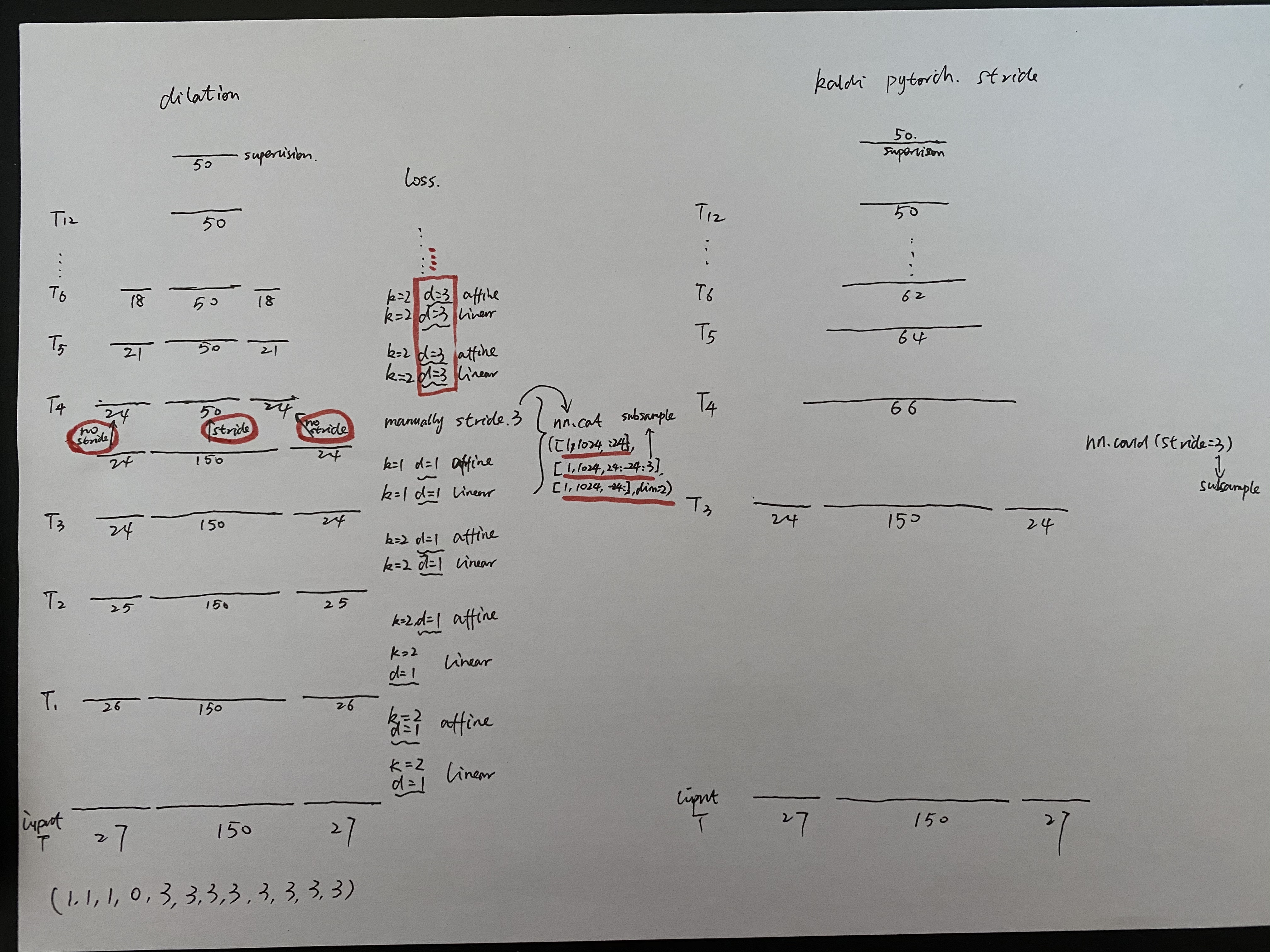
Just subsample the t_out_length from (24+150+24) to (24+50+24) manually, the number of parameters will not increase than stride kernel(2,2) version. I explained this code behaviour in the picture below.
| self.conv = nn.Conv1d(in_channels=dim, | ||
| out_channels=bottleneck_dim, | ||
| kernel_size=kernel_size, | ||
| dilation=dilation, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
You should never need the dilation parameter. I think we discussed this before.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
... instead of using dilation, do a 3-fold subsampling after the last layer that had stride=1. Please don't argue about this! I remember last time was quite painful.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
hah, I find the discussed info before. I just to make the length of output is equal to the supervision
2020-02-11 14:59:29,145 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 202])
2020-02-11 14:59:29,150 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 200])
2020-02-11 14:59:29,156 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 198])
2020-02-11 14:59:29,162 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 98])
2020-02-11 14:59:29,175 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 92])
2020-02-11 14:59:29,184 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 86])
2020-02-11 14:59:29,195 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 80])
2020-02-11 14:59:29,204 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 74])
2020-02-11 14:59:29,215 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 68])
2020-02-11 14:59:29,224 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 62])
2020-02-11 14:59:29,233 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 56])
2020-02-11 14:59:29,243 (model_tdnnf3:172) DEBUG: x shape is torch.Size([1, 1024, 50])
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Are these shape of output generated by tdnnf layers correct?
2020-02-11 19:21:40,284 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 202])
2020-02-11 19:21:40,291 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 200])
2020-02-11 19:21:40,297 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 198])
2020-02-11 19:21:40,306 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 198])
2020-02-11 19:21:40,317 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 192])
2020-02-11 19:21:40,327 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 186])
2020-02-11 19:21:40,336 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 180])
2020-02-11 19:21:40,346 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 174])
2020-02-11 19:21:40,355 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 168])
2020-02-11 19:21:40,364 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 162])
2020-02-11 19:21:40,373 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 156])
2020-02-11 19:21:40,382 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 150])
2020-02-11 19:21:40,382 (model_tdnnf3:182) DEBUG: x shape is torch.Size([1, 1024, 50])
|
I think lufan was not involved last time and he doesn't know this.
Sorry I only have time from tomorrow.
Sent from myMail for iOS
Tuesday, 11 February 2020, 18:36 +0800 from [email protected] <[email protected]>:
…
@danpovey commented on this pull request.
----------------------------------------------------------------------
In egs/aishell/s10/chain/tdnnf_layer.py :
>
# conv requires [N, C, T]
self.conv = nn.Conv1d(in_channels=dim,
out_channels=bottleneck_dim,
kernel_size=kernel_size,
+ dilation=dilation,
You should never need the dilation parameter. I think we discussed this before.
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub , or unsubscribe .
|
|
I will help you tomorrow.
Sent from myMail for iOS
Tuesday, 11 February 2020, 19:04 +0800 from [email protected] <[email protected]>:
…
@fanlu commented on this pull request.
----------------------------------------------------------------------
In egs/aishell/s10/chain/model.py :
> @@ -174,6 +170,13 @@ def forward(self, x):
# tdnnf requires input of shape [N, C, T]
for i in range(len(self.tdnnfs)):
x = self.tdnnfs[i](x)
+ # stride manually, do not stride context
+ if self.tdnnfs[i].time_stride == 0:
+ cur_context = sum(self.time_stride_list[i:])
+ x_left = x[:, :, :cur_context]
+ x_mid = x[:, :, cur_context:-cur_context:self.frame_subsampling_factor]
+ x_right = x[:, :, -cur_context:]
+ x = torch.cat([x_left, x_mid, x_right], dim=2)
I want subsample the length of window only rather than left_context and right_context. And this is slower than before training, but it worked. please help me to write this 1d convolution.
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub , or unsubscribe .
|
thanks |
|
Not really: somewhere near the beginning, there should be a subsampling by
a factor of 3. The original script was much closer to being correct, it
only needed a very small change.
…On Tue, Feb 11, 2020 at 7:22 PM fanlu ***@***.***> wrote:
***@***.**** commented on this pull request.
------------------------------
In egs/aishell/s10/chain/tdnnf_layer.py
<#3923 (comment)>:
>
# conv requires [N, C, T]
self.conv = nn.Conv1d(in_channels=dim,
out_channels=bottleneck_dim,
kernel_size=kernel_size,
+ dilation=dilation,
Are these shape of output generated by tdnnf layers correct?
2020-02-11 19:21:40,284 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 202])
2020-02-11 19:21:40,291 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 200])
2020-02-11 19:21:40,297 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 198])
2020-02-11 19:21:40,306 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 198])
2020-02-11 19:21:40,317 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 192])
2020-02-11 19:21:40,327 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 186])
2020-02-11 19:21:40,336 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 180])
2020-02-11 19:21:40,346 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 174])
2020-02-11 19:21:40,355 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 168])
2020-02-11 19:21:40,364 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 162])
2020-02-11 19:21:40,373 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 156])
2020-02-11 19:21:40,382 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 150])
2020-02-11 19:21:40,382 (model_tdnnf3:182) DEBUG: x shape is torch.Size([1, 1024, 50])
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#3923?email_source=notifications&email_token=AAZFLO4CMV65SVJAOGDDNSTRCKDA7A5CNFSM4KS576D2YY3PNVWWK3TUL52HS4DFWFIHK3DMKJSXC5LFON2FEZLWNFSXPKTDN5WW2ZLOORPWSZGOCVAPXHQ#discussion_r377575919>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AAZFLO22IBPTMVDRSDCUNMTRCKDA7ANCNFSM4KS576DQ>
.
|
|
... i.e. make the bottleneck-dim of the prefinal layer a separate parameter
(set it to 256), and set affine=False in all the batchnorm modules.
…On Tue, Feb 11, 2020 at 7:29 PM Daniel Povey ***@***.***> wrote:
Not really: somewhere near the beginning, there should be a subsampling by
a factor of 3. The original script was much closer to being correct, it
only needed a very small change.
On Tue, Feb 11, 2020 at 7:22 PM fanlu ***@***.***> wrote:
> ***@***.**** commented on this pull request.
> ------------------------------
>
> In egs/aishell/s10/chain/tdnnf_layer.py
> <#3923 (comment)>:
>
> >
> # conv requires [N, C, T]
> self.conv = nn.Conv1d(in_channels=dim,
> out_channels=bottleneck_dim,
> kernel_size=kernel_size,
> + dilation=dilation,
>
> Are these shape of output generated by tdnnf layers correct?
>
> 2020-02-11 19:21:40,284 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 202])
> 2020-02-11 19:21:40,291 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 200])
> 2020-02-11 19:21:40,297 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 198])
> 2020-02-11 19:21:40,306 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 198])
> 2020-02-11 19:21:40,317 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 192])
> 2020-02-11 19:21:40,327 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 186])
> 2020-02-11 19:21:40,336 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 180])
> 2020-02-11 19:21:40,346 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 174])
> 2020-02-11 19:21:40,355 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 168])
> 2020-02-11 19:21:40,364 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 162])
> 2020-02-11 19:21:40,373 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 156])
> 2020-02-11 19:21:40,382 (model_tdnnf3:179) DEBUG: x shape is torch.Size([1, 1024, 150])
> 2020-02-11 19:21:40,382 (model_tdnnf3:182) DEBUG: x shape is torch.Size([1, 1024, 50])
>
> —
> You are receiving this because you were mentioned.
> Reply to this email directly, view it on GitHub
> <#3923?email_source=notifications&email_token=AAZFLO4CMV65SVJAOGDDNSTRCKDA7A5CNFSM4KS576D2YY3PNVWWK3TUL52HS4DFWFIHK3DMKJSXC5LFON2FEZLWNFSXPKTDN5WW2ZLOORPWSZGOCVAPXHQ#discussion_r377575919>,
> or unsubscribe
> <https://github.com/notifications/unsubscribe-auth/AAZFLO22IBPTMVDRSDCUNMTRCKDA7ANCNFSM4KS576DQ>
> .
>
|
OK, I will try the original script again with affine=False in bn and prefinal layer with 256. |
|
I have changed the dilation to stride and kernel_size of (linear,affine) from (3,1) to (2,2)
|
|
You can format the table to the format supported by Markdown and Github will render it correctly. |
|
I have test this two version on speed pertub data with hires feature and tree leaves from 5k to 4k corresponding tdnn_1c.
|
|
Regarding the stride vs. dilation difference, you would have to show me
some kind of diff.
Perhaps a PR to your own repo?
If done right, they would be equivalent; I'd have to look.
…On Wed, Feb 12, 2020 at 9:42 AM fanlu ***@***.***> wrote:
I have test this two version on speed pertub data with hires feature and
tree leaves from 5k to 4k corresponding tdnn_1c.
Here is the result
stride kernel(2,2) dilation kernel(2,2)
dev_cer 7.06 6.50
dev_wer 15.18 14.52
test_cer 8.57 7.81
test_wer 17.33 16.42
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#3923?email_source=notifications&email_token=AAZFLOYHVL6L2YV2LARYQL3RCNHXLA5CNFSM4KS576D2YY3PNVWWK3TUL52HS4DFVREXG43VMVBW63LNMVXHJKTDN5WW2ZLOORPWSZGOELPAETA#issuecomment-584974924>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AAZFLO7LLUXS3PR5HSZ2CY3RCNHXLANCNFSM4KS576DQ>
.
|
|
no stride with left and right context may be give the model more context info to make model more robust with test result |
|
That doesn't sound right to me. Please show me the diff.
…On Wed, Feb 12, 2020 at 10:48 AM fanlu ***@***.***> wrote:
no stride with left and right context may be give the model more context
info to make model more robust with test result
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#3923?email_source=notifications&email_token=AAZFLOYPZFPOZWX64U5AW2LRCNPQFA5CNFSM4KS576D2YY3PNVWWK3TUL52HS4DFVREXG43VMVBW63LNMVXHJKTDN5WW2ZLOORPWSZGOELPETMQ#issuecomment-584993202>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AAZFLO3EE3LS6CG6YTH6TX3RCNPQFANCNFSM4KS576DQ>
.
|
| kernel_size=1) | ||
| self.batchnorm2 = nn.BatchNorm1d(num_features=small_dim) | ||
|
|
||
| def forward(self, x): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm surprised you didn't implement the TDNN_F layer in the "obvious" way with 1-d convolution.
[oh, sorry, this is prefinal layer]
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Inside OrthonormalLinear, it is nn.Conv1d. So it is ineeded implemented via 1-d convolution.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
kernel_size=1 doesn't look right. Some extremely weird stuff is going on in this PR.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hi, dan, this code's behavior is not different with before code's. just changed the param. the original stride version use kernel_size=1 as default also
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
It's getting less clear to me, not more clear, and in any case the code is not right.
You showed various experimental numbers but you never said with any clarity what code or parameters each one corresponded to.
I would prefer if you just went back to the original code and made the small and very specific changes that I asked for.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
sorry about this
|
sorry, dan, I am confused about diff. Do I have to do anything else?
|
You showed a table with columns:
|
Noticed that the dilation version must be different with original kaldi version. |
|
@fanlu Regarding the
|
|
Compared the speed of this two version
|
|
Yes. I have noticed that. So I called this is a new version.
|
|
But never mind, the result of |
OK, there are a couple problems with this. I'm confident that's what you are doing in the middle layer (with time_stride=0 in Kaldi) is not right, but what's going on is a bit strange and I'm not quite sure what it's doing, partly because I'm not sure where that code goes. But it doesn't matter.. let me describe how I think it should be done. There issue of clarity, in that the time_stride is very unintuitive in this context. Better EDIT: Later we can figure out why in your experiments stride was not faster than dilation-- it definitely should be faster, as the flops differ by nearly a factor of 3. There must have been some problem or bug or unexpected machine load. |
|
Thanks, Dan, Let me try to implement this based on #3925
|
|
.. also, I suspect that in your "dilated" version of the code, the layer with (in Kaldi terminology) time_stride=0 had effectively time_stride=1. That may be why the WER differed. |
|
sorry about mismatch
|
which is correct?
|
|
@danpovey this is the pr of stride kernel(2,2) https://github.com/mobvoi/kaldi/pull/1, please have a look |
|
OK.
That's not right though, I'm afraid.
(I mean, the code is doing something super strange that can't at all be the
right thing).
…On Wed, Feb 12, 2020 at 9:22 PM fanlu ***@***.***> wrote:
***@***.**** commented on this pull request.
------------------------------
In egs/aishell/s10/chain/model.py
<#3923 (comment)>:
> @@ -174,6 +170,13 @@ def forward(self, x):
# tdnnf requires input of shape [N, C, T]
for i in range(len(self.tdnnfs)):
x = self.tdnnfs[i](x)
+ # stride manually, do not stride context
+ if self.tdnnfs[i].time_stride == 0:
+ cur_context = sum(self.time_stride_list[i:])
+ x_left = x[:, :, :cur_context]
+ x_mid = x[:, :, cur_context:-cur_context:self.frame_subsampling_factor]
+ x_right = x[:, :, -cur_context:]
+ x = torch.cat([x_left, x_mid, x_right], dim=2)
Just subsample the t_out_length from (24+150+24) to (24+50+24) manually,
the number of parameters will not increase than *stride kernel(2,2)*
version. I explained this code behaviour in the picture below.
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#3923?email_source=notifications&email_token=AAZFLO5QFOWTYNDLCHYZLZLRCPZXZA5CNFSM4KS576D2YY3PNVWWK3TUL52HS4DFWFIHK3DMKJSXC5LFON2FEZLWNFSXPKTDN5WW2ZLOORPWSZGOCVHCY6Y#discussion_r378245403>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AAZFLO7KK2F6XKI26GJMRRDRCPZXZANCNFSM4KS576DQ>
.
|
|
considering the kaldi version. the left or right context is
|
|
I am assuming this is not ready to merge, since I am still seeing |
|
I will close it since of #3925 |
Hi, @danpovey @csukuangfj. please review this tdnnf version. thanks