|
391 | 391 |
|
392 | 392 | <br /> |
393 | 393 |
|
394 | | -* Overview of the neuroimaging pipeline structure |
395 | | - |
396 | 394 | * Why do data require preprocessing? |
397 | 395 |
|
| 396 | +* Overview of the neuroimaging pipeline structure |
| 397 | + |
398 | 398 | * *fMRIPrep* |
399 | 399 |
|
400 | 400 | * [Hi]story behind the tool |
|
410 | 410 |
|
411 | 411 | --- |
412 | 412 |
|
| 413 | +## fMRI Preprocessing Recap |
| 414 | + |
| 415 | +.boxed-content[ |
| 416 | + |
| 417 | +.large[ |
| 418 | + |
| 419 | +Goal: Ensure the time series at each voxel is meaningful |
| 420 | + |
| 421 | +* Drawn from the same location at all time points |
| 422 | + * Motion correction |
| 423 | +* Location can be described anatomically |
| 424 | + * Coregistration, susceptibility distortion correction |
| 425 | +* Location can be selected by standard coordinates |
| 426 | + * Spatial normalization |
| 427 | + |
| 428 | +The quality of analysis is limited by the quality of preprocessing. |
| 429 | + |
| 430 | +] |
| 431 | + |
| 432 | +] |
| 433 | + |
| 434 | +--- |
| 435 | + |
| 436 | +## Reproducibility |
| 437 | + |
| 438 | + |
| 439 | +.center[ |
| 440 | +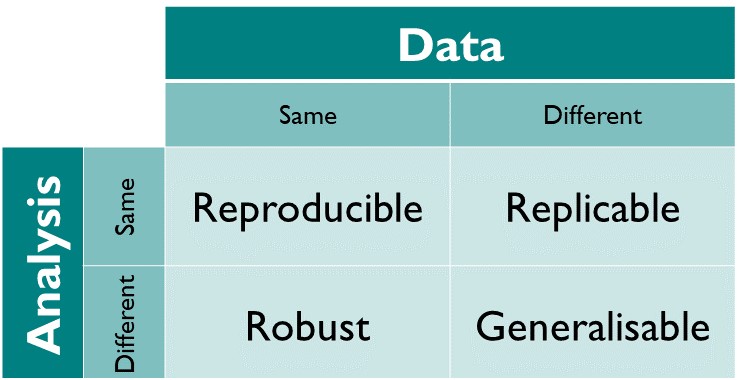 |
| 441 | +.pull-right[(from [The Turing Way](https://book.the-turing-way.org/reproducible-research/overview/overview-definitions#table-of-definitions-for-reproducibility))] |
| 442 | +] |
| 443 | + |
| 444 | +.boxed-content[ |
| 445 | + |
| 446 | +.large[ |
| 447 | + |
| 448 | +The reproducibility of analysis is limited by the reproducibility of preprocessing. |
| 449 | + |
| 450 | +] |
| 451 | + |
| 452 | +] |
| 453 | + |
| 454 | +--- |
| 455 | + |
413 | 456 | ## The neuroimaging worflow |
414 | 457 |
|
415 | 458 | <br /> |
|
433 | 476 |
|
434 | 477 | --- |
435 | 478 |
|
436 | | -template: title |
437 | | -layout: false |
| 479 | +## fMRIPrep |
| 480 | + |
| 481 | +.boxed-content[ |
| 482 | + |
| 483 | +.large[ |
| 484 | + |
| 485 | +Robust, generic fMRI preprocessing |
| 486 | +* Works with the data you give it, imposing minimal requirements |
| 487 | + * You can run it on a T1w scan and a BOLD series |
| 488 | + * Will utilize additional scans (T2w/FLAIR, field maps) when available |
| 489 | + |
| 490 | +Cross-suite tooling |
| 491 | +* Select tools from each suite according to strengths |
| 492 | +* Use Nipype to abstract from tool details and construct modular workflows |
| 493 | + |
| 494 | +Standardized |
| 495 | +* NIfTI, JSON and TSV inputs (BIDS) |
| 496 | +* NIfTI, GIFTI, CIFTI-2, JSON and TSV outputs (BIDS Derivatives) |
| 497 | + |
| 498 | +] |
| 499 | + |
| 500 | +] |
| 501 | + |
| 502 | +--- |
| 503 | + |
| 504 | +## fMRIPrep |
| 505 | + |
438 | 506 | .middle[ |
439 | | -<p align="center"> |
| 507 | +<p align="right"> |
440 | 508 | <img src="https://github.com/oesteban/fmriprep/raw/f4c7a9804be26c912b24ef4dccba54bdd72fa1fd/docs/_static/fmriprep-21.0.0.svg" width="95%" /> |
441 | 509 | </p> |
442 | 510 | ] |
|
0 commit comments