|
492 | 492 | .pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> |
493 | 493 | ] |
494 | 494 | ] |
| 495 | + |
| 496 | +--- |
| 497 | + |
495 | 498 | .right-column2.larger[ |
496 | 499 |
|
497 | 500 | * BIDS is a directory structure, based on common practices |
|
525 | 528 | ]] |
526 | 529 |
|
527 | 530 | --- |
528 | | -name: newsection |
| 531 | +template: newsection |
529 | 532 | layout: true |
530 | 533 |
|
531 | | -.perma-sidebar[ |
532 | | -<p class="rotate"> |
533 | | -<a rel="license" href="http://creativecommons.org/licenses/by/4.0/"><img alt="Creative Commons License" style="border-width:0; height: 20px; padding-top: 6px;" src="https://i.creativecommons.org/l/by/4.0/88x31.png" /></a> |
534 | | - <span style="padding-left: 10px; font-weight: 600;">Day 1 :: Brain Imaging Data Structure</span> |
535 | | -</p> |
536 | | -] |
537 | 534 | --- |
538 | 535 |
|
539 | 536 | # Converting from DICOM into BIDS |
|
653 | 650 |
|
654 | 651 | --- |
655 | 652 |
|
656 | | -# Bidsification: ReproIn |
| 653 | +# DICOM-to-BIDS: ReproIn |
| 654 | + |
| 655 | +.center[[ReproIn](https://github.com/ReproNim/reproin) standardizes scanner conventions:] |
657 | 656 |
|
658 | | -The [ReproIn](https://github.com/ReproNim/reproin) project aims to |
659 | | -standardize scanner conventions, to eliminate the need to rewrite these mappings. |
| 657 | +<figure style="width: 60%"> |
| 658 | +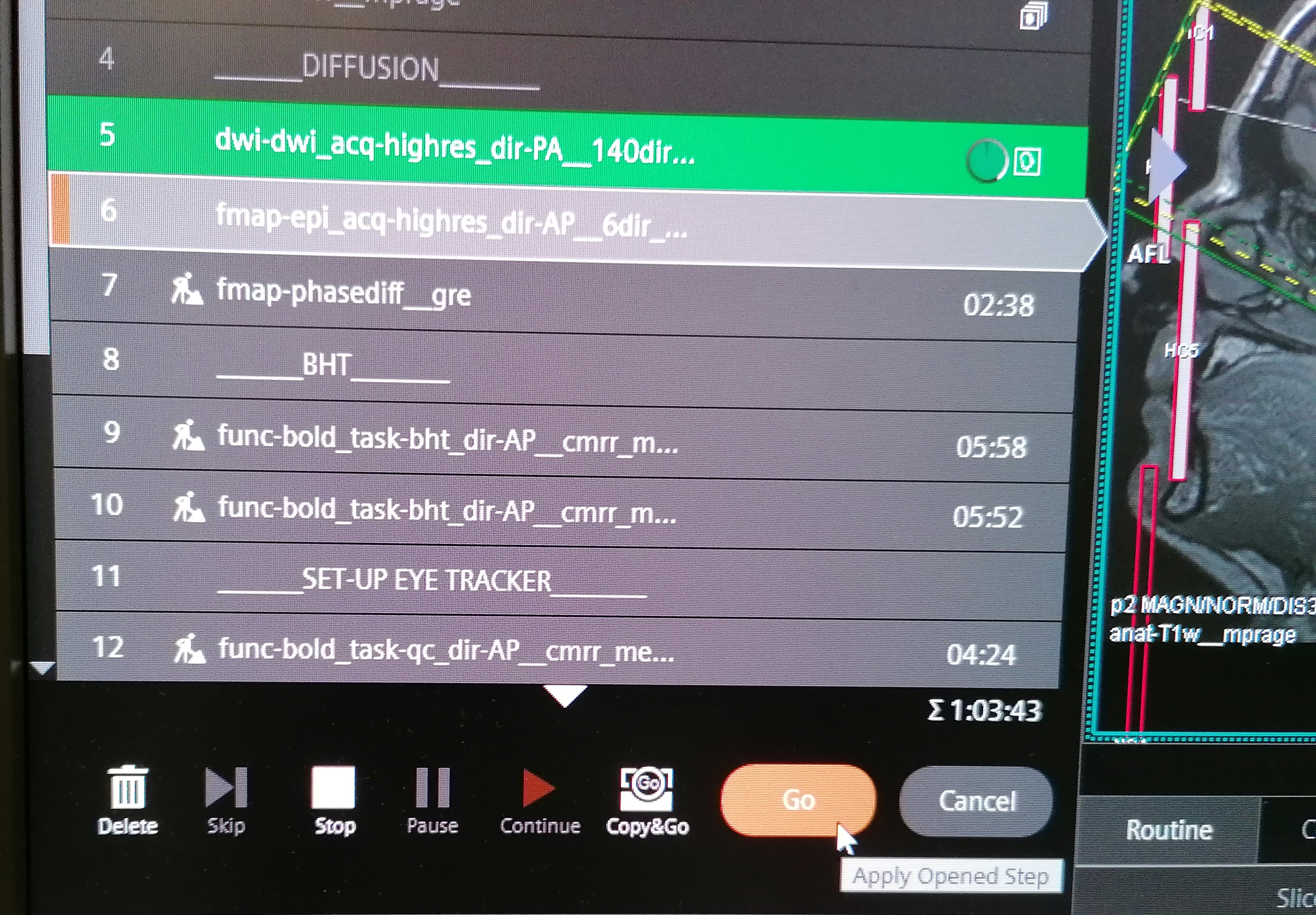 |
| 659 | +<figcaption>From <a href="https://www.axonlab.org/hcph-sops">the HCPh SOPs</a>; |
| 660 | +doi:</figcaption> |
| 661 | +</figure> |
| 662 | + |
| 663 | +--- |
660 | 664 |
|
661 | | -<figure style="width: 40%"> |
| 665 | +<figure style="width: 60%"> |
662 | 666 |  |
663 | 667 | <figcaption>From <a href="https://github.com/ReproNim/reproin/blob/master/README.md#overall-workflow">ReproIn</a>; |
664 | 668 | doi:</figcaption> |
665 | | -</figure> |
| 669 | + |
| 670 | +??? |
| 671 | + |
| 672 | +Please note that this figure is OUT OF DATE. For example, the anatomicals are called anat-T1W__, anat-T2w__ etc. |
| 673 | + |
| 674 | +--- |
| 675 | + |
| 676 | +# DICOM-to-BIDS: HeuDiConv |
| 677 | + |
| 678 | +.boxed-content.no-bullet[ |
| 679 | + |
| 680 | +* .large[<i class="fa-solid fa-circle-chevron-right"></i> **Heu**ristic **DI**COM** Conv**erter: maps DICOM metadata into BIDS] |
| 681 | + |
| 682 | +* .large[<i class="fa-solid fa-circle-chevron-right"></i> Mappings are encoded within the *heuristic file*] |
| 683 | + |
| 684 | + * The `.heudiconv/` folder contains the metadata that can be employed in the *heuristic file* to define mappings.<br /><br /> |
| 685 | + |
| 686 | +<figure style="width: 100%"> |
| 687 | +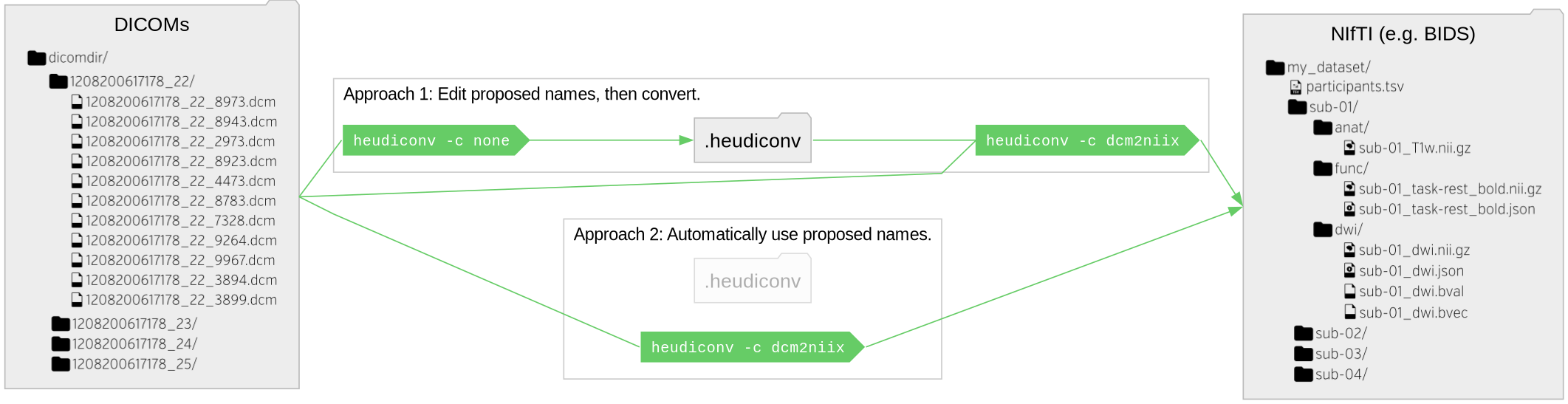 |
| 688 | +<figcaption>From <a href="https://github.com/nipy/heudiconv?tab=readme-ov-file#howto-101">HeuDiConv</a>; |
| 689 | +doi:</figcaption> |
| 690 | + |
| 691 | +] |
| 692 | + |
| 693 | +--- |
| 694 | + |
| 695 | +# DICOM-to-BIDS: HeuDiConv |
| 696 | + |
| 697 | +.boxed-content[ |
| 698 | + |
| 699 | +.no-bullet.large[ |
| 700 | +* <i class="fa-solid fa-1"></i> Generate a **heuristic file** |
| 701 | + |
| 702 | +* <i class="fa-solid fa-2"></i> Edit the **heuristic file** |
| 703 | +] |
| 704 | + |
| 705 | +.small[ |
| 706 | +```Python |
| 707 | +data = create_key('run-{item:03d}') |
| 708 | + |
| 709 | +t1w = create_key('sub-{subject}/{session}/anat/sub-{subject}_{session}_T1w') |
| 710 | + |
| 711 | +dwi = create_key('sub-{subject}/{session}/dwi/sub-{subject}_{session}_dir-AP_dwi') |
| 712 | + |
| 713 | +# Save the RPE (reverse phase-encode) B0 image as a fieldmap (fmap). It will be used to correct |
| 714 | +# the distortion in the DWI |
| 715 | +fmap_rev_phase = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_dir-PA_epi') |
| 716 | + |
| 717 | +fmap_mag = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_magnitude') |
| 718 | + |
| 719 | +fmap_phase = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_phasediff') |
| 720 | + |
| 721 | +# Even if this is resting state, you still need a task key |
| 722 | +func_rest = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-rest_run-01_bold') |
| 723 | +func_rest_post = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-rest_run-02_bold') |
| 724 | +``` |
| 725 | +] |
| 726 | + |
| 727 | +.no-bullet.large[ |
| 728 | +* <i class="fa-solid fa-3"></i> Execute *HeuDiConv* |
| 729 | +] |
| 730 | + |
| 731 | +] |
| 732 | + |
| 733 | +--- |
| 734 | + |
| 735 | +# DICOM-to-BIDS: ReproIn + HeuDiConv |
| 736 | + |
| 737 | +.no-bullet.large[ |
| 738 | +* <i class="fa-solid fa-circle-chevron-right"></i> Just run *HeuDiConv* directly! *Heuristic file* is pre-defined. |
| 739 | + |
| 740 | +* <i class="fa-solid fa-circle-question"></i> What if I **did not** follow *ReproIn* namings? |
| 741 | +] |
| 742 | + |
| 743 | +-- |
| 744 | + |
| 745 | +.boxed-content.small[ |
| 746 | + |
| 747 | +``` Python |
| 748 | +# A dictionary containing fixes/remapping for sequence names per study. |
| 749 | +# Keys are md5sum of study_description from DICOMs, in the form of PI-Experimenter^protocolname |
| 750 | +# You can use `heudiconv -f reproin --command ls --files PATH |
| 751 | +# to list the "study hash". |
| 752 | +# Values are list of tuples in the form (regex_pattern, substitution). |
| 753 | +# If the key is an empty string "", it would apply to any study. |
| 754 | +protocols2fix: dict[str | re.Pattern[str], list[tuple[str, str]]] = { |
| 755 | + "": [ # <-- empty string means apply to whole study |
| 756 | + ("anat-T1w_acq-mp2rage_run-01", "anat-mp2rage"), |
| 757 | + ("_UNI_", "_acq-mp2rage__"), |
| 758 | + ("_UNI-DEN", "_acq-denoised__"), |
| 759 | + ("_INV", "_inv-"), |
| 760 | + ("_T1_", "_acq-T1map__"), |
| 761 | + ("fmap_acq-siemens", "fmap-phasediff"), |
| 762 | + ("fmap-gre_acq-siemens", "fmap-phasediff"), |
| 763 | + ("_acq-midRes", ""), |
| 764 | + ("_acq-p6", ""), |
| 765 | + ("_run-01", ""), |
| 766 | + ] |
| 767 | +} |
| 768 | +``` |
| 769 | + |
| 770 | +] |
| 771 | + |
| 772 | +--- |
| 773 | + |
| 774 | +# DICOM-to-BIDS: ReproIn + HeuDiConv |
| 775 | + |
| 776 | +.left-column-mid[ |
| 777 | +Our dataset's DICOM had some non-compliant names: |
| 778 | +.small[ |
| 779 | +.pad-left[ |
| 780 | +<i class="fa-solid fa-folder-open"></i> my_dataset/<br /> |
| 781 | +.pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> |
| 782 | +.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 10 - anat-T1w_acq-mp2rage_run-01<mark>_UNI_Images</mark>.dicom]]<br /> |
| 783 | +.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 11 - anat-T1w_acq-mp2rage_run-01<mark>_UNI-DEN</mark>.dicom]]<br /> |
| 784 | +.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 12 - anat-T1w_acq-mp2rage_run-01<mark>_INV2</mark>.dicom]]<br /> |
| 785 | +.pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 16 - fmap-gre_acq-siemens_run-01/]]<br /> |
| 786 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> |
| 787 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> |
| 788 | +.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 17 - <mark>fmap-gre</mark>_acq-siemens_run-01.dicom ]]<br /> |
| 789 | +.pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 18 - func-bold_acq-midRes_task-rest_run-01/ ]]<br /> |
| 790 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> |
| 791 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-ellipsis"></i>]]]<br /> |
| 792 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> |
| 793 | +.pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 20 - func-bold_acq-midRes_task-mixed_run-01/ ]]<br /> |
| 794 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> |
| 795 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-ellipsis"></i>]]]<br /> |
| 796 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> |
| 797 | +.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 22 - anat-T2w_acq-p6_run-01.dicom]]<br /> |
| 798 | +.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 7 - anat-T1w_acq-mp2rage_run-01<mark>_INV1</mark>.dicom]]<br /> |
| 799 | +.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 8 - anat-T1w_acq-mp2rage_run-01<mark>_T1_Images</mark>.dicom]]<br /> |
| 800 | +.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 9 - anat-T1w_acq-mp2rage_run-01<mark>_T1_Images</mark>.dicom]]<br /> |
| 801 | +.pad-left[<i class="fa-solid fa-folder"></i> sub-14]<br /> |
| 802 | +.pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> |
| 803 | +] |
| 804 | +]] |
| 805 | + |
| 806 | +.right-column-mid.small[ |
| 807 | +.pad-left[ |
| 808 | +<i class="fa-solid fa-folder-open"></i> my_dataset/<br /> |
| 809 | +.pad-left[<i class="fa-solid fa-file-lines"></i> CHANGES]<br /> |
| 810 | +.pad-left[<i class="fa-solid fa-file-lines"></i> README]<br /> |
| 811 | +.pad-left[<i class="fa-solid fa-paperclip"></i> dataset_description.json]<br /> |
| 812 | +.pad-left[<i class="fa-solid fa-paperclip"></i> participants.json]<br /> |
| 813 | +.pad-left[<i class="fa-solid fa-table"></i> participants.tsv]<br /> |
| 814 | +.pad-left[<i class="fa-solid fa-paperclip"></i> scans.json]<br /> |
| 815 | +.pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> |
| 816 | +.pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> anat/]]<br /> |
| 817 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_T2w.json]]]<br /> |
| 818 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_T2w.nii.gz]]]<br /> |
| 819 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_acq-denoised_T1w.json]]]<br /> |
| 820 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-denoised_T1w.nii.gz]]]<br /> |
| 821 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_acq-mp2rage_T1w.json]]]<br /> |
| 822 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-mp2rage_T1w.nii.gz]]]<br /> |
| 823 | +.pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> fmap/]]<br /> |
| 824 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_magnitude1.json]]]<br /> |
| 825 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_magnitude1.nii.gz]]]<br /> |
| 826 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_magnitude2.json]]]<br /> |
| 827 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_magnitude2.nii.gz]]]<br /> |
| 828 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_phasediff.json]]]<br /> |
| 829 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_phasediff.nii.gz]]]<br /> |
| 830 | +.pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> func/]]<br /> |
| 831 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-mixed_bold.json]]]<br /> |
| 832 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-mixed_bold.nii.gz]]]<br /> |
| 833 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-table"></i> sub-11_task-mixed_events.tsv]]]<br /> |
| 834 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-rest_bold.json]]]<br /> |
| 835 | +.pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-rest_bold.nii.gz]]]<br /> |
| 836 | +.pad-left[.pad-left[<i class="fa-solid fa-table"></i> sub-11_scans.tsv]]<br /> |
| 837 | +.pad-left[<i class="fa-solid fa-folder"></i> sub-14]<br /> |
| 838 | +.pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> |
| 839 | +] |
| 840 | +] |
| 841 | + |
| 842 | +<i class="fa-solid fa-circle-right" style="margin-top: 20%; font-size: 1.7em"></i> |
666 | 843 |
|
667 | 844 | --- |
668 | 845 |
|
|
0 commit comments