diff --git a/README.md b/README.md
index 434881e..11ef2fc 100644
--- a/README.md
+++ b/README.md
@@ -11,6 +11,33 @@ This combined repository includes tutorials and code resources provided by the N
## Tutorials
+### [ICESat-2_Cloud_Access Notebooks](./notebooks/ICESat-2_Cloud_Access)
+
+These notebooks demonstrate how to search and access ICESat-2 from the NASA Earthdata Cloud:
+
+**Accessing and working with ICESat-2 Data in the Cloud**
+
+Originally presented to the UWG (User Working Group) in May 2022, this tutorial demonstrates how to search for ICESat-2 data hosted in the Earthdata Cloud and how to directly access it from an Amazon Web Services (AWS) Elastic Compute Cloud (EC2) instance using the `earthaccess` package.
+
+**Plotting ICESat-2 and CryoSat-2 Freeboards**
+
+This notebook demonstrates plotting ICESat-2 and CryoSat-2 data in the same map from within an AWS ec2 instance. ICESat-2 data are accessed via "direct S3 access" using `earthaccess`. CryoSat-2 data are downloaded to our cloud instance from their ftp storage lcoation and accessed locally.
+
+**Processing Large-scale Time Series of ICESat-2 Sea Ice Height in the Cloud**
+
+This notebook utilizes several libraries to performantly search, access, read, and grid ATL10 data over the Ross Sea, Antarctica including `earthaccess`, `h5coro`, and `geopandas`. The notebook provides further guidance on how to scale this analysis to the entire continent, running the same workflow from a script that can be run from your laptop using [Coiled](https://www.coiled.io/).
+
+### [MEaSUREs](./notebooks/measures)
+
+**Download, crop, resample, and plot multiple GeoTIFFs**
+
+This tutorial guides you through programmatically accessing and downloading GeoTIFF files from the NSIDC DAAC to your local computer. We then crop and resample one GeoTIFF based on the extent and pixel size of another GeoTIFF, then plot one on top of the other.
+
+We will use two data sets from the NASA [MEaSUREs](https://nsidc.org/data/measures) (Making Earth System data records for Use in Research Environments) program as an example:
+
+* [MEaSUREs Greenland Ice Mapping Project (GrIMP) Digital Elevation Model from GeoEye and WorldView Imagery, Version 2 (NSIDC-0715)](https://nsidc.org/data/nsidc-0715/versions/2)
+* [MEaSUREs Greenland Ice Velocity: Selected Glacier Site Velocity Maps from InSAR, Version 4 (NSIDC-0481)](https://nsidc.org/data/nsidc-0481/versions/4)
+
### [SnowEx_ASO_MODIS_Snow](./notebooks/SnowEx_ASO_MODIS_Snow)
**Snow Depth and Snow Cover Data Exploration**
@@ -23,6 +50,13 @@ Originally demonstrated through the NASA Earthdata Webinar "Let It Snow! Accessi
Originally presented during the 2019 AGU Fall Meeting, this tutorial demonstrates the NSIDC DAAC's data discovery, access, and subsetting services, along with basic open source resources used to harmonize and analyze data across multiple products. The tutorial is provided as a series of Python-based Jupyter Notebooks, focusing on sea ice height and ice surface temperature data from NASA’s ICESat-2 and MODIS missions, respectively, to characterize Arctic sea ice.
+### [ITS_LIVE](./notebooks/itslive)
+
+**Global land ice velocities.**
+The Inter-mission Time Series of Land Ice Velocity and Elevation (ITS_LIVE) project facilitates ice sheet, ice shelf and glacier research by providing a globally comprehensive and temporally dense multi-sensor record of land ice velocity and elevation with low latency. Scene-pair velocities were generated from satellite optical and radar imagery.
+
+The notebooks on this project demonstrate how to search and access ITS_LIVE velocity pairs and provide a simple example on how to build a data cube.
+
### [IceFlow](./notebooks/iceflow)
> [!CAUTION]
@@ -37,31 +71,6 @@ Originally presented during the 2019 AGU Fall Meeting, this tutorial demonstrate
**Harmonized data for pre-IceBridge, ICESat and IceBridge data sets.**
These Jupyter notebooks are interactive documents to teach students and researchers interested in cryospheric sciences how to access and work with airborne altimetry and related data sets from NASA’s [IceBridge](https://www.nasa.gov/mission_pages/icebridge/index.html) mission, and satellite altimetry data from [ICESat](https://icesat.gsfc.nasa.gov/icesat/) and [ICESat-2](https://icesat-2.gsfc.nasa.gov/) missions using the NSIDC **IceFlow API**
-
-### [ITS_LIVE](./notebooks/itslive)
-
-**Global land ice velocities.**
-The Inter-mission Time Series of Land Ice Velocity and Elevation (ITS_LIVE) project facilitates ice sheet, ice shelf and glacier research by providing a globally comprehensive and temporally dense multi-sensor record of land ice velocity and elevation with low latency. Scene-pair velocities were generated from satellite optical and radar imagery.
-
-The notebooks on this project demonstrate how to search and access ITS_LIVE velocity pairs and provide a simple example on how to build a data cube.
-
-### [ICESat-2_Cloud_Access](./notebooks/ICESat-2_Cloud_Access)
-
-**Accessing and working with ICESat-2 Data in the Cloud**
-
-Originally presented to the UWG (User Working Group) in May 2022, this tutorial demonstrates how to search for ICESat-2 data hosted in the Earthdata Cloud and how to directly access it from an Amazon Web Services (AWS) Elastic Compute Cloud (EC2) instance using the `earthaccess` package.
-
-### [MEaSUREs](./notebooks/measures)
-
-**Download, crop, resample, and plot multiple GeoTIFFs**
-
-This tutorial guides you through programmatically accessing and downloading GeoTIFF files from the NSIDC DAAC to your local computer. We then crop and resample one GeoTIFF based on the extent and pixel size of another GeoTIFF, then plot one on top of the other.
-
-We will use two data sets from the NASA [MEaSUREs](https://nsidc.org/data/measures) (Making Earth System data records for Use in Research Environments) program as an example:
-
-* [MEaSUREs Greenland Ice Mapping Project (GrIMP) Digital Elevation Model from GeoEye and WorldView Imagery, Version 2 (NSIDC-0715)](https://nsidc.org/data/nsidc-0715/versions/2)
-* [MEaSUREs Greenland Ice Velocity: Selected Glacier Site Velocity Maps from InSAR, Version 4 (NSIDC-0481)](https://nsidc.org/data/nsidc-0481/versions/4)
-
## Usage with Binder
The Binder button above allows you to explore and run the notebook in a shared cloud computing environment without the need to install dependencies on your local machine. Note that this option will not directly download data to your computer; instead the data will be downloaded to the cloud environment.
diff --git a/notebooks/ICESat-2_Cloud_Access/ATL06-direct-access.ipynb b/notebooks/ICESat-2_Cloud_Access/ATL06-direct-access.ipynb
index 0d65230..0ec39f6 100644
--- a/notebooks/ICESat-2_Cloud_Access/ATL06-direct-access.ipynb
+++ b/notebooks/ICESat-2_Cloud_Access/ATL06-direct-access.ipynb
@@ -2,18 +2,18 @@
"cells": [
{
"cell_type": "markdown",
- "id": "c91169d7",
+ "id": "e0754304-7036-4530-83ec-86cec0f9886b",
"metadata": {
"tags": []
},
"source": [
- "\n",
- " \n",
- " \n",
+ "\n",
+ "\n",
"# **Accessing and working with ICESat-2 data in the cloud**\n",
- " \n",
- "\n",
- " \n",
+ "\n",
+ "\n",
+ "\n",
+ " \n",
"\n",
"## **1. Tutorial Overview**\n",
"\n",
@@ -44,18 +44,15 @@
"\n",
"### **Prerequisites**\n",
"\n",
- "1. An Amazon Web Services (AWS) account. You will be accessing data in Amazon Web Services (AWS) Simple Storage Service (S3) buckets. This requires an AWS account, for details on how to set one up see [here](https://nsidc.org/data/user-resources/help-center/nasa-earthdata-cloud-data-access-guide#anchor-1).\n",
- "2. An EC2 instance in the us-west-2 region is set up. **NASA cloud-hosted data is in Amazon Region us-west2. So you also need an EC2 instance in the us-west-2 region.** An EC2 instance is a virtual computer that you create to perform processing operations in place of using your own desktop or laptop. Details on how to set up an instance can be found [here](https://nsidc.org/data/user-resources/help-center/nasa-earthdata-cloud-data-access-guide#anchor-1).\n",
- "3. An Earthdata Login is required for data access. If you don't have one, you can register for one [here](https://urs.earthdata.nasa.gov/).\n",
- "4. A .netrc file, that contains your Earthdata Login credentials, in your home directory. The current recommended practice for authentication is to create a .netrc file in your home directory following [these instructions](https://nsidc.org/support/how/how-do-i-programmatically-request-data-services) (Step 1) and to use the .netrc file for authentication when required for data access during the tutorial.\n",
- "5. The *nsidc-tutorials* environment is setup and activated. This [README](https://github.com/nsidc/NSIDC-Data-Tutorials/blob/main/README.md) has setup instructions.\n",
+ "1. An EC2 instance in the us-west-2 region. **NASA cloud-hosted data is in Amazon Region us-west2. So you also need an EC2 instance in the us-west-2 region.** An EC2 instance is a virtual computer that you create to perform processing operations in place of using your own desktop or laptop. Details on how to set up an instance can be found [here](https://nsidc.org/data/user-resources/help-center/nasa-earthdata-cloud-data-access-guide#anchor-1).\n",
+ "2. An Earthdata Login is required for data access. If you don't have one, you can register for one [here](https://urs.earthdata.nasa.gov/).\n",
+ "3. A .netrc file, that contains your Earthdata Login credentials, in your home directory. The current recommended practice for authentication is to create a .netrc file in your home directory following [these instructions](https://nsidc.org/support/how/how-do-i-programmatically-request-data-services) (Step 1) and to use the .netrc file for authentication when required for data access during the tutorial.\n",
+ "4. The *nsidc-tutorials* environment is setup and activated. This [README](https://github.com/nsidc/NSIDC-Data-Tutorials/blob/main/README.md) has setup instructions.\n",
"\n",
"### **Example of end product** \n",
"At the end of this tutorial, the following figure will be generated:\n",
- "\n",
- "
\n",
- " \n",
+ "\n",
+ "\n",
"# **Accessing and working with ICESat-2 data in the cloud**\n",
- " \n",
- "\n",
- " \n",
+ "\n",
+ "\n",
+ "\n",
+ " \n",
"\n",
"## **1. Tutorial Overview**\n",
"\n",
@@ -44,18 +44,15 @@
"\n",
"### **Prerequisites**\n",
"\n",
- "1. An Amazon Web Services (AWS) account. You will be accessing data in Amazon Web Services (AWS) Simple Storage Service (S3) buckets. This requires an AWS account, for details on how to set one up see [here](https://nsidc.org/data/user-resources/help-center/nasa-earthdata-cloud-data-access-guide#anchor-1).\n",
- "2. An EC2 instance in the us-west-2 region is set up. **NASA cloud-hosted data is in Amazon Region us-west2. So you also need an EC2 instance in the us-west-2 region.** An EC2 instance is a virtual computer that you create to perform processing operations in place of using your own desktop or laptop. Details on how to set up an instance can be found [here](https://nsidc.org/data/user-resources/help-center/nasa-earthdata-cloud-data-access-guide#anchor-1).\n",
- "3. An Earthdata Login is required for data access. If you don't have one, you can register for one [here](https://urs.earthdata.nasa.gov/).\n",
- "4. A .netrc file, that contains your Earthdata Login credentials, in your home directory. The current recommended practice for authentication is to create a .netrc file in your home directory following [these instructions](https://nsidc.org/support/how/how-do-i-programmatically-request-data-services) (Step 1) and to use the .netrc file for authentication when required for data access during the tutorial.\n",
- "5. The *nsidc-tutorials* environment is setup and activated. This [README](https://github.com/nsidc/NSIDC-Data-Tutorials/blob/main/README.md) has setup instructions.\n",
+ "1. An EC2 instance in the us-west-2 region. **NASA cloud-hosted data is in Amazon Region us-west2. So you also need an EC2 instance in the us-west-2 region.** An EC2 instance is a virtual computer that you create to perform processing operations in place of using your own desktop or laptop. Details on how to set up an instance can be found [here](https://nsidc.org/data/user-resources/help-center/nasa-earthdata-cloud-data-access-guide#anchor-1).\n",
+ "2. An Earthdata Login is required for data access. If you don't have one, you can register for one [here](https://urs.earthdata.nasa.gov/).\n",
+ "3. A .netrc file, that contains your Earthdata Login credentials, in your home directory. The current recommended practice for authentication is to create a .netrc file in your home directory following [these instructions](https://nsidc.org/support/how/how-do-i-programmatically-request-data-services) (Step 1) and to use the .netrc file for authentication when required for data access during the tutorial.\n",
+ "4. The *nsidc-tutorials* environment is setup and activated. This [README](https://github.com/nsidc/NSIDC-Data-Tutorials/blob/main/README.md) has setup instructions.\n",
"\n",
"### **Example of end product** \n",
"At the end of this tutorial, the following figure will be generated:\n",
- "\n",
- " \n",
- "\n",
- "\n",
+ " \n",
+ "\n",
"### **Time requirement**\n",
"\n",
"Allow approximately 20 minutes to complete this tutorial."
@@ -397,7 +394,7 @@
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
- "version": "3.9.15"
+ "version": "3.10.14"
}
},
"nbformat": 4,
diff --git a/notebooks/ICESat-2_Cloud_Access/ICESat2-CryoSat2.ipynb b/notebooks/ICESat-2_Cloud_Access/ICESat2-CryoSat2.ipynb
new file mode 100644
index 0000000..b08d9b3
--- /dev/null
+++ b/notebooks/ICESat-2_Cloud_Access/ICESat2-CryoSat2.ipynb
@@ -0,0 +1,478 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "id": "de81ea4e-1dad-4f1d-af4b-416dd256af6f",
+ "metadata": {},
+ "source": [
+ "# **Plotting ICESat-2 and CryoSat-2 Freeboards**\n",
+ "\n",
+ "\n",
+ "\n",
+ "\n",
+ "### **Credits**\n",
+ "This notebook was created by Mikala Beig and Andy Barrett, NSIDC\n",
+ "\n",
+ "### **Learning Objectives** \n",
+ "\n",
+ "1. use `earthaccess` to search for ICESat-2 ATL10 data using a spatial filter\n",
+ "2. open cloud-hosted files using direct access to the ICESat-2 S3 bucket; \n",
+ "3. use cs2eo script to download files into your hub instance\n",
+ "3. load an HDF5 group into an `xarray.Dataset`; \n",
+ "4. visualize freeboards using `hvplot`.\n",
+ "5. map the locations of ICESat-2 and CryoSat-2 freeboards using `cartopy`\n",
+ "\n",
+ "### **Prerequisites**\n",
+ "\n",
+ "1. An EC2 instance in the us-west-2 region. **NASA cloud-hosted data are in Amazon Region us-west2. So you also need an EC2 instance in the us-west-2 region.** .\n",
+ "2. An Earthdata Login is required for data access. If you don't have one, you can register for one [here](https://urs.earthdata.nasa.gov/).\n",
+ "3. Experience using cs2eo to query for coincident data.\n",
+ "4. A cs2eo download script for CryoSat-2 data.\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "a2db2b3c-97bf-42aa-8fd1-0eb588afa80e",
+ "metadata": {},
+ "source": [
+ "### **Tutorial Steps**\n",
+ "\n",
+ "#### Query for coincident ICESat-2 and CryoSat-2 data\n",
+ "\n",
+ "Using the cs2eo coincident data explorer, query for ATL10 and CryoSat-2, L2, SAR, POCA, Baseline E data products using a spatial and temporal filter.\n",
+ "\n",
+ "**Download the basic result metadata and the raw access scripts.** Upload the ESA download script (SIR_SAR_L2_E_download_script.py) into the folder from which you are running this notebook.\n",
+ "\n",
+ ""
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "94cbed67-e58a-4579-b153-c9fcbe1e2ab4",
+ "metadata": {},
+ "source": [
+ "#### Import Packages"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e911afb1-b247-4412-9342-1e8865b3084e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "import os\n",
+ "import platform\n",
+ "from ftplib import FTP\n",
+ "import sys\n",
+ "\n",
+ "\n",
+ "# For searching and accessing NASA data\n",
+ "import earthaccess\n",
+ "\n",
+ "# For reading data, analysis and plotting\n",
+ "import xarray as xr\n",
+ "import hvplot.xarray\n",
+ "\n",
+ "# For nice printing of python objects\n",
+ "import pprint \n",
+ "\n",
+ "# For plotting\n",
+ "import matplotlib.pyplot as plt\n",
+ "import cartopy.crs as ccrs\n",
+ "import cartopy.feature as cfeature\n",
+ "\n",
+ "#downloading files using cs2eo script\n",
+ "from SIR_SAR_L2_E_download_script import download_files\n",
+ "\n",
+ "## use your own email here\n",
+ "user_email = 'your email here'\n",
+ "path = './data/'"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "58c3ef53-7223-42da-afd4-e14e87caa85d",
+ "metadata": {},
+ "source": [
+ "#### Download CryoSat-2 data to your hub instance\n",
+ "\n",
+ "Copy the list of ESA files from within SIR_SAR_L2_E_download_script.py "
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "ac8e451e-b687-4c13-a5b8-9b700a519a46",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "esa_files = ['SIR_SAR_L2/2019/12/CS_LTA__SIR_SAR_2__20191227T110305_20191227T111751_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200329T163208_20200329T164044_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200114T203033_20200114T204440_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191103T134759_20191103T135125_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200204T191657_20200204T192558_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/12/CS_LTA__SIR_SAR_2__20191216T215645_20191216T220909_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200315T065755_20200315T071241_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191030T135252_20191030T135600_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200219T081800_20200219T083303_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200110T203717_20200110T204612_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200409T053748_20200409T054151_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200413T053254_20200413T053659_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200208T191154_20200208T192117_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200319T065300_20200319T070802_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200304T175209_20200304T180102_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191128T122800_20191128T123212_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191009T150801_20191009T151142_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191121T231659_20191121T232817_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200215T082253_20200215T083741_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200121T094259_20200121T095800_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191005T151255_20191005T151621_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200427T150701_20200427T151544_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191024T004201_20191024T005059_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200308T174708_20200308T175621_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200402T162707_20200402T163602_E001.nc']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "eaa73f90-84f0-4429-854a-784c95afde66",
+ "metadata": {},
+ "source": [
+ "Download the CryoSat-2 files into your hub instance by calling the download_files function you imported from the script."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "ecef7122-b3d5-4ca0-b207-7ad3c35ca34d",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "download_files(user_email, esa_files)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6e9d27c7-d9d9-40ae-aab6-34888aedfe8f",
+ "metadata": {},
+ "source": [
+ "Stashing the files in a data folder to keep our notebook directory less cluttered."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2f6c0779-77cc-4d60-a7a6-cb4a0ce1ea25",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "!mv CS_LTA__SIR*.nc data"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "aeb468e5-7a5a-456c-96cb-6808af3f5eaa",
+ "metadata": {},
+ "source": [
+ "#### Use `earthaccess` for querying and direct S3 access of ATL10\n",
+ "\n",
+ "First we authenticate using `earthaccess`"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2165876a-c5fa-4df9-b7f5-c82f01e7bdba",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "auth = earthaccess.login()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "fb7adbf2-1148-4075-8578-91e01fbeacfc",
+ "metadata": {},
+ "source": [
+ "Then we use a spatial filter to search for ATL10 granules that intersect our area of interest. This is the same area we used in our cs2eo query above."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "37139d13-43e4-4563-b436-2879c76677ec",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "results = earthaccess.search_data(\n",
+ " short_name = 'ATL10',\n",
+ " version = '006',\n",
+ " cloud_hosted = True,\n",
+ " bounding_box = (-17, 79, 12, 83),\n",
+ " temporal = ('2019-10-01','2020-04-30'),\n",
+ " count = 10\n",
+ ")\n",
+ "#note that with count=10 we're limiting the number of files we actually access for this tutorial."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "b883fc7f-1c26-471c-88c4-664fa59bed16",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "display(results[1])"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e58e1111-1b07-47ec-952b-036290346f4c",
+ "metadata": {},
+ "source": [
+ "We use earthaccess.open() to directly access the ATL10 files within their S3 bucket. earthaccess.open() creates a file-like object, which is required because AWS S3 uses object storage, and we need to create a virtual file system to work with the HDF5 library."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "a80212ab-ee66-4d06-8a4a-47169092159e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "%time\n",
+ "icesat2_files = earthaccess.open(results) \n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "d24f9d50-3f95-4cdf-b877-1ac06be4471b",
+ "metadata": {},
+ "source": [
+ "We can use xarray to examine the contents of our files (one group at a time)."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "1d1102e5-561e-41e8-81df-e4ff543d19c1",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds_is2 = xr.open_dataset(icesat2_files[1], group='gt2r/freeboard_segment')\n",
+ "ds_is2"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "c603f6a6-7dcb-4a6f-9130-0bab933ff3c6",
+ "metadata": {},
+ "source": [
+ "And we can use hvplot to plot one of the variables within that group."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "0503dfee-7d66-49fa-8482-ad2dc330ff52",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds_is2['beam_fb_height'].hvplot(kind='scatter', s=2)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6a89c207-028f-47df-b7d9-5723bd5e836f",
+ "metadata": {},
+ "source": [
+ "#### Open and plot downloaded CryoSat-2 data \n",
+ "\n",
+ "We need a list of the downloaded CryoSat-2 files."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "60c1704a-0b17-4d4f-a7a8-dac8fe5b9701",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "downloaded_files = os.listdir(path)\n",
+ "downloaded_files"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "2d280de0-5de7-4a0c-9986-c94ab6b6d05a",
+ "metadata": {},
+ "source": [
+ "We use xarray to access the contents of our netcdf file. In this case, we are not \"streaming\" data from an S3 bucket, but are accessing the data locally."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "8657ed36-d583-46b9-8ed2-9b1b57cb5786",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds_cs2 = xr.open_dataset(path + downloaded_files[0])\n",
+ "ds_cs2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "795b0bdd-0895-4de4-8008-b6f15fcf2b3e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds_cs2['radar_freeboard_20_ku'].hvplot(kind='scatter', s=2)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "40a8164d-f5c1-4049-aefe-aeb08295092b",
+ "metadata": {},
+ "source": [
+ "#### Plot ICESat-2 and CryoSat-2 Freeboards on same map"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "ad5f7f2c-2ff8-47cc-8ea8-337f9183c20e",
+ "metadata": {},
+ "source": [
+ "Here we're plotting one file from each data set to save time."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "11476d13-0f1f-44f7-a364-4a949554d8c2",
+ "metadata": {},
+ "outputs": [],
+ "source": [
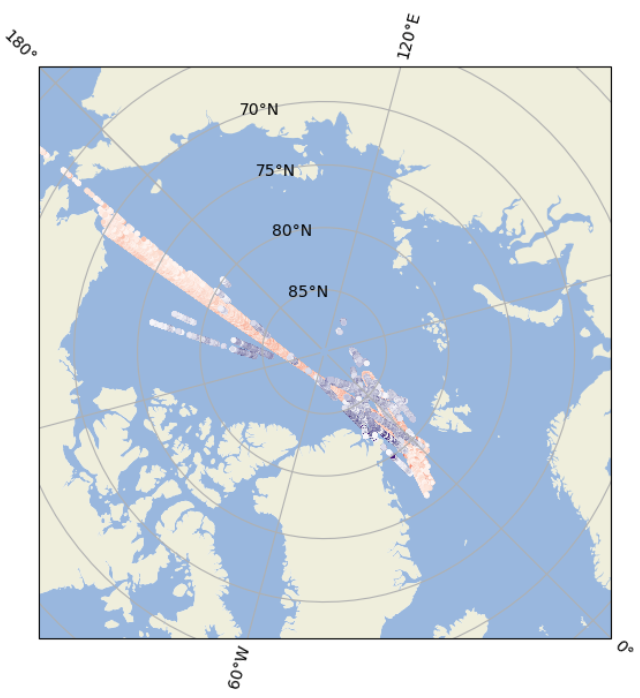
+ "projection = ccrs.Stereographic(central_latitude=90.,\n",
+ " central_longitude=-45.,\n",
+ " true_scale_latitude=70.)\n",
+ "extent = [-2500000.000, 2500000., -2500000., 2500000.000]\n",
+ "\n",
+ "\n",
+ "fig = plt.figure(figsize=(10,10))\n",
+ "ax = fig.add_subplot(projection=projection)\n",
+ "ax.set_extent(extent, projection)\n",
+ "ax.add_feature(cfeature.OCEAN)\n",
+ "ax.add_feature(cfeature.LAND)\n",
+ "ax.gridlines(draw_labels=True)\n",
+ "\n",
+ "vmin = 0.\n",
+ "vmax = 1.\n",
+ "\n",
+ "# Plot Cryosat freeboard\n",
+ "cs2_img = ax.scatter(ds_cs2.lon_poca_20_ku, ds_cs2.lat_poca_20_ku, 5,\n",
+ " c=ds_cs2.radar_freeboard_20_ku, \n",
+ " vmin=vmin, vmax=vmax, # Set max and min values for plotting\n",
+ " cmap='Reds', # shading='auto' to avoid warning\n",
+ " transform=ccrs.PlateCarree()) # coords are lat,lon but map if NPS \n",
+ "\n",
+ "# Plot IS2 freeboard \n",
+ "is2_img = ax.scatter(ds_is2.longitude, ds_is2.latitude, 5,\n",
+ " c=ds_is2.beam_fb_height, \n",
+ " vmin=vmin, vmax=vmax, \n",
+ " cmap='Purples', \n",
+ " transform=ccrs.PlateCarree())\n",
+ "\n",
+ "# Add colorbars\n",
+ "fig.colorbar(cs2_img, label='Cryosat-2 Radar Freeboard (m)')\n",
+ "fig.colorbar(is2_img, label='ICESat-2 Lidar Freeboard (m)')"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "392bb8fd-8ec6-4fd4-b6db-dc0386094c7f",
+ "metadata": {},
+ "source": [
+ "Here we're plotting several ICESat-2 and CryoSat-2 files at a time. This takes a few minutes to render."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "4815c13b-88c3-4d10-850e-02cf766d1844",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "%%time\n",
+ "\n",
+ "# NSIDC WGS84 Polar Stereographic \n",
+ "projection = ccrs.Stereographic(central_latitude=90.,\n",
+ " central_longitude=-45.,\n",
+ " true_scale_latitude=70.)\n",
+ "extent = [-2500000.000, 2500000., -2500000., 2500000.000]\n",
+ "\n",
+ "fig = plt.figure(figsize=(10,10))\n",
+ "ax = fig.add_subplot(projection=projection)\n",
+ "ax.set_extent(extent, projection)\n",
+ "# ax.coastlines()\n",
+ "ax.add_feature(cfeature.OCEAN)\n",
+ "ax.add_feature(cfeature.LAND)\n",
+ "\n",
+ "ax.gridlines(draw_labels=True)\n",
+ "\n",
+ "vmin = 0.\n",
+ "vmax = 1.\n",
+ "\n",
+ "# Plot CryoSat-2 freeboards\n",
+ "for fp in downloaded_files:\n",
+ " ds = xr.open_dataset(path + fp)\n",
+ " cs2= plt.scatter(ds.lon_poca_20_ku, ds.lat_poca_20_ku, 5,\n",
+ " c=ds.radar_freeboard_20_ku, cmap=\"Reds\",\n",
+ " vmin=vmin, vmax=vmax,\n",
+ " transform=ccrs.PlateCarree())\n",
+ "\n",
+ "# Plot ICESat-2 freeboards\n",
+ "for fp in icesat2_files:\n",
+ " ds = xr.open_dataset(fp, group='gt2r/freeboard_segment')\n",
+ " is2 = plt.scatter(ds.longitude, ds.latitude, 5,\n",
+ " c=ds.beam_fb_height, cmap=\"Purples\",\n",
+ " vmin=vmin, vmax=vmax,\n",
+ " transform=ccrs.PlateCarree())\n",
+ " \n",
+ "fig.colorbar(cs2, label=\"CryoSat-2 Radar Freeboard (m)\")\n",
+ "fig.colorbar(is2, label=\"ICESat-2 Lidar Freeboard (m)\")\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "a4681430-14b4-4a97-9a0a-9f3f587dfb8a",
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "Python 3 (ipykernel)",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.10.14"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/notebooks/ICESat-2_Cloud_Access/README.md b/notebooks/ICESat-2_Cloud_Access/README.md
index c95904b..b77b3fd 100644
--- a/notebooks/ICESat-2_Cloud_Access/README.md
+++ b/notebooks/ICESat-2_Cloud_Access/README.md
@@ -1,24 +1,50 @@
# ICESat-2 Cloud Access
## Summary
-This notebook demonstrates searching for cloud-hosted ICESat-2 data and directly accessing Land Ice Height (ATL06) granules from an Amazon Compute Cloud (EC2) instance using the `earthaccess` package. NASA data "in the cloud" are stored in Amazon Web Services (AWS) Simple Storage Service (S3) Buckets. **Direct Access** is an efficient way to work with data stored in an S3 Bucket when you are working in the cloud. Cloud-hosted granules can be opened and loaded into memory without the need to download them first. This allows you take advantage of the scalability and power of cloud computing.
+We provide several notebooks showcasing how to search and access ICESat-2 from the NASA Earthdata Cloud. NASA data "in the cloud" are stored in Amazon Web Services (AWS) Simple Storage Service (S3) Buckets. **Direct Access** is an efficient way to work with data stored in an S3 Bucket when you are working in the cloud. Cloud-hosted granules can be opened and loaded into memory without the need to download them first. This allows you take advantage of the scalability and power of cloud computing.
+
+### [Accessing and working with ICESat-2 data in the cloud](./ATL06-direct-access_rendered.ipynb)
+This notebook demonstrates searching for cloud-hosted ICESat-2 data and directly accessing Land Ice Height (ATL06) granules from an Amazon Compute Cloud (EC2) instance using the `earthaccess` package.
+
+#### Key Learning Objectives
+1. Use `earthaccess` to search for ICESat-2 data using spatial and temporal filters and explore search results;
+2. Open data granules using direct access to the ICESat-2 S3 bucket;
+3. Load a HDF5 group into an `xarray.Dataset`;
+4. Visualize the land ice heights using `hvplot`.
+
+### [Plotting ICESat-2 and CryoSat-2 Freeboards](./ICESat2-CryoSat2.ipynb)
+This notebook demonstrates plotting ICESat-2 and CryoSat-2 data in the same map from within an AWS ec2 instance. ICESat-2 data are accessed via "direct S3 access" using `earthaccess`. CryoSat-2 data are downloaded to our cloud instance from their ftp storage lcoation and accessed locally.
+
+#### Key Learning Objectives
+1. use `earthaccess` to search for ICESat-2 ATL10 data using a spatial filter
+2. open cloud-hosted files using direct access to the ICESat-2 S3 bucket;
+3. use cs2eo script to download files into your hub instance
+3. load an HDF5 group into an `xarray.Dataset`;
+4. visualize freeboards using `hvplot`.
+5. map the locations of ICESat-2 and CryoSat-2 freeboards using `cartopy`
+
+### [Processing Large-scale Time Series of ICESat-2 Sea Ice Height in the Cloud](./ATL10-h5coro_rendered.ipynb)
+This notebook utilizes several libraries to performantly search, access, read, and grid ATL10 data over the Ross Sea, Antarctica including `earthaccess`, `h5coro`, and `geopandas`. The notebook provides further guidance on how to scale this analysis to the entire continent, running the same workflow from a script that can be run from your laptop using [Coiled](https://www.coiled.io/).
+
+#### Key Learning Objectives
+1. Use earthaccess to authenticate with Earthdata Login, search for ICESat-2 data using spatial and temporal filters, and directly access files in the cloud.
+2. Open data granules using h5coro to efficiently read HDF5 data from the NSIDC DAAC S3 bucket.
+3. Load data into a geopandas.DataFrame containing geodetic coordinates, ancillary variables, and date/time converted from ATLAS Epoch.
+4. Grid track data to EASE-Grid v2 6.25 km projected grid using drop-in-the-bucket resampling.
+5. Calculate mean statistics and assign aggregated data to grid cells.
+6. Visualize aggregated sea ice height data on a map.
## Set up
-To run the notebook provided in this folder in the Amazon Web Services (AWS) cloud, there are a couple of options:
-* An EC2 instance already set up with the necessary software installed to run a Jupyter notebook, and the environment set up using the provided environment.yml file. **Note:** If you are running this notebook on your own AWS EC2 instance using the environment set up using the environment.yml file in the NSIDC-Data-Tutorials/notebooks/ICESat-2_Cloud_Access/environment folder, you may need to run the following command before running the notebook to ensure the notebook executes properly:
+To run the notebooks provided in this folder in the Amazon Web Services (AWS) cloud, there are a couple of options:
+* An EC2 instance already set up with the necessary software installed to run a Jupyter notebook, and the environment set up using the provided environment.yml file. **Note:** If you are running these notebooks on your own AWS EC2 instance using the environment set up using the environment.yml file in the NSIDC-Data-Tutorials/notebooks/ICESat-2_Cloud_Access/environment folder, you may need to run the following command before running the notebook to ensure the notebook executes properly:
`jupyter nbextension enable --py widgetsnbextension`
You do NOT need to do this if you are using the environment set up using the environment.yml file from the NSIDC-Data-Tutorials/binder folder.
-* Alternatively, if you have access to one, it can be run in a managed cloud-based Jupyter hub. Just make sure all the necessary libraries are installed (`earthaccess`,`xarray`, and `hvplot`).
+* Alternatively, if you have access to one, it can be run in a managed cloud-based Jupyter hub. Just make sure all the necessary libraries are installed (e.g. `earthaccess`,`xarray`,`hvplot`, etc.).
-For further details on the prerequisites, see the 'Prerequisites' section in the notebook.
+For further details on the prerequisites, see the 'Prerequisites' section in each notebook.
-## Key Learning Objectives
-1. Use `earthaccess` to search for ICESat-2 data using spatial and temporal filters and explore search results;
-2. Open data granules using direct access to the ICESat-2 S3 bucket;
-3. Load a HDF5 group into an `xarray.Dataset`;
-4. Visualize the land ice heights using `hvplot`.
diff --git a/notebooks/ICESat-2_Cloud_Access/SIR_SAR_L2_E_download_script.py b/notebooks/ICESat-2_Cloud_Access/SIR_SAR_L2_E_download_script.py
new file mode 100644
index 0000000..a4d564a
--- /dev/null
+++ b/notebooks/ICESat-2_Cloud_Access/SIR_SAR_L2_E_download_script.py
@@ -0,0 +1,63 @@
+import os
+import platform
+from ftplib import FTP
+import sys
+
+
+download_file_obj = None
+read_byte_count = None
+total_byte_count = None
+
+
+def get_padded_count(count, max_count):
+ return str(count).zfill(len(str(max_count)))
+
+
+def file_byte_handler(data):
+ global download_file_obj, read_byte_count, total_byte_count
+ download_file_obj.write(data)
+ read_byte_count = read_byte_count + len(data)
+ progress_bar(read_byte_count, total_byte_count)
+
+
+def progress_bar(progress, total, prefix="", size=60, file=sys.stdout):
+ if total != 0:
+ x = int(size * progress / total)
+ x_percent = int(100 * progress / total)
+ file.write(f" {prefix} [{'='*x}{' '*(size-x)}] {x_percent} % \r")
+ file.flush()
+
+
+def download_files(user_email, esa_files):
+ global download_file_obj, read_byte_count, total_byte_count
+ print("About to connect to ESA science server")
+ with FTP("science-pds.cryosat.esa.int") as ftp:
+ try:
+ ftp.login("anonymous", user_email)
+ print("Downloading {} files".format(len(esa_files)))
+
+ for i, filename in enumerate(esa_files):
+ padded_count = get_padded_count(i + 1, len(esa_files))
+ print("{}/{}. Downloading file {}".format(padded_count, len(esa_files), os.path.basename(filename)))
+
+ with open(os.path.basename(filename), 'wb') as download_file:
+ download_file_obj = download_file
+ total_byte_count = ftp.size(filename)
+ read_byte_count = 0
+ ftp.retrbinary('RETR ' + filename, file_byte_handler, 1024)
+ print("\n")
+ finally:
+ print("Exiting FTP.")
+ ftp.quit()
+
+
+if __name__ == '__main__':
+
+ esa_files = ['SIR_SAR_L2/2019/12/CS_LTA__SIR_SAR_2__20191227T110305_20191227T111751_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200329T163208_20200329T164044_E001.nc', 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200114T203033_20200114T204440_E001.nc', 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191103T134759_20191103T135125_E001.nc', 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200204T191657_20200204T192558_E001.nc', 'SIR_SAR_L2/2019/12/CS_LTA__SIR_SAR_2__20191216T215645_20191216T220909_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200315T065755_20200315T071241_E001.nc', 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191030T135252_20191030T135600_E001.nc', 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200219T081800_20200219T083303_E001.nc', 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200110T203717_20200110T204612_E001.nc', 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200409T053748_20200409T054151_E001.nc', 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200413T053254_20200413T053659_E001.nc', 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200208T191154_20200208T192117_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200319T065300_20200319T070802_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200304T175209_20200304T180102_E001.nc', 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191128T122800_20191128T123212_E001.nc', 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191009T150801_20191009T151142_E001.nc', 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191121T231659_20191121T232817_E001.nc', 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200215T082253_20200215T083741_E001.nc', 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200121T094259_20200121T095800_E001.nc', 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191005T151255_20191005T151621_E001.nc', 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200427T150701_20200427T151544_E001.nc', 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191024T004201_20191024T005059_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200308T174708_20200308T175621_E001.nc', 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200402T162707_20200402T163602_E001.nc']
+
+ if int(platform.python_version_tuple()[0]) < 3:
+ exit("Your Python version is {}. Please use version 3.0 or higher.".format(platform.python_version()))
+
+ email = input("Please enter your e-mail: ")
+
+ download_files(email, esa_files)
diff --git a/notebooks/ICESat-2_Cloud_Access/img/ATL10_CS2_L2_SAR_query_med.png b/notebooks/ICESat-2_Cloud_Access/img/ATL10_CS2_L2_SAR_query_med.png
new file mode 100644
index 0000000..1579b12
Binary files /dev/null and b/notebooks/ICESat-2_Cloud_Access/img/ATL10_CS2_L2_SAR_query_med.png differ
diff --git a/notebooks/ICESat-2_Cloud_Access/img/icesat2-cryosat2.png b/notebooks/ICESat-2_Cloud_Access/img/icesat2-cryosat2.png
new file mode 100644
index 0000000..b2ca77d
Binary files /dev/null and b/notebooks/ICESat-2_Cloud_Access/img/icesat2-cryosat2.png differ
\n",
- "\n",
- "\n",
+ " \n",
+ "\n",
"### **Time requirement**\n",
"\n",
"Allow approximately 20 minutes to complete this tutorial."
@@ -397,7 +394,7 @@
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
- "version": "3.9.15"
+ "version": "3.10.14"
}
},
"nbformat": 4,
diff --git a/notebooks/ICESat-2_Cloud_Access/ICESat2-CryoSat2.ipynb b/notebooks/ICESat-2_Cloud_Access/ICESat2-CryoSat2.ipynb
new file mode 100644
index 0000000..b08d9b3
--- /dev/null
+++ b/notebooks/ICESat-2_Cloud_Access/ICESat2-CryoSat2.ipynb
@@ -0,0 +1,478 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "id": "de81ea4e-1dad-4f1d-af4b-416dd256af6f",
+ "metadata": {},
+ "source": [
+ "# **Plotting ICESat-2 and CryoSat-2 Freeboards**\n",
+ "\n",
+ "\n",
+ "\n",
+ "\n",
+ "### **Credits**\n",
+ "This notebook was created by Mikala Beig and Andy Barrett, NSIDC\n",
+ "\n",
+ "### **Learning Objectives** \n",
+ "\n",
+ "1. use `earthaccess` to search for ICESat-2 ATL10 data using a spatial filter\n",
+ "2. open cloud-hosted files using direct access to the ICESat-2 S3 bucket; \n",
+ "3. use cs2eo script to download files into your hub instance\n",
+ "3. load an HDF5 group into an `xarray.Dataset`; \n",
+ "4. visualize freeboards using `hvplot`.\n",
+ "5. map the locations of ICESat-2 and CryoSat-2 freeboards using `cartopy`\n",
+ "\n",
+ "### **Prerequisites**\n",
+ "\n",
+ "1. An EC2 instance in the us-west-2 region. **NASA cloud-hosted data are in Amazon Region us-west2. So you also need an EC2 instance in the us-west-2 region.** .\n",
+ "2. An Earthdata Login is required for data access. If you don't have one, you can register for one [here](https://urs.earthdata.nasa.gov/).\n",
+ "3. Experience using cs2eo to query for coincident data.\n",
+ "4. A cs2eo download script for CryoSat-2 data.\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "a2db2b3c-97bf-42aa-8fd1-0eb588afa80e",
+ "metadata": {},
+ "source": [
+ "### **Tutorial Steps**\n",
+ "\n",
+ "#### Query for coincident ICESat-2 and CryoSat-2 data\n",
+ "\n",
+ "Using the cs2eo coincident data explorer, query for ATL10 and CryoSat-2, L2, SAR, POCA, Baseline E data products using a spatial and temporal filter.\n",
+ "\n",
+ "**Download the basic result metadata and the raw access scripts.** Upload the ESA download script (SIR_SAR_L2_E_download_script.py) into the folder from which you are running this notebook.\n",
+ "\n",
+ ""
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "94cbed67-e58a-4579-b153-c9fcbe1e2ab4",
+ "metadata": {},
+ "source": [
+ "#### Import Packages"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e911afb1-b247-4412-9342-1e8865b3084e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "import os\n",
+ "import platform\n",
+ "from ftplib import FTP\n",
+ "import sys\n",
+ "\n",
+ "\n",
+ "# For searching and accessing NASA data\n",
+ "import earthaccess\n",
+ "\n",
+ "# For reading data, analysis and plotting\n",
+ "import xarray as xr\n",
+ "import hvplot.xarray\n",
+ "\n",
+ "# For nice printing of python objects\n",
+ "import pprint \n",
+ "\n",
+ "# For plotting\n",
+ "import matplotlib.pyplot as plt\n",
+ "import cartopy.crs as ccrs\n",
+ "import cartopy.feature as cfeature\n",
+ "\n",
+ "#downloading files using cs2eo script\n",
+ "from SIR_SAR_L2_E_download_script import download_files\n",
+ "\n",
+ "## use your own email here\n",
+ "user_email = 'your email here'\n",
+ "path = './data/'"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "58c3ef53-7223-42da-afd4-e14e87caa85d",
+ "metadata": {},
+ "source": [
+ "#### Download CryoSat-2 data to your hub instance\n",
+ "\n",
+ "Copy the list of ESA files from within SIR_SAR_L2_E_download_script.py "
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "ac8e451e-b687-4c13-a5b8-9b700a519a46",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "esa_files = ['SIR_SAR_L2/2019/12/CS_LTA__SIR_SAR_2__20191227T110305_20191227T111751_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200329T163208_20200329T164044_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200114T203033_20200114T204440_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191103T134759_20191103T135125_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200204T191657_20200204T192558_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/12/CS_LTA__SIR_SAR_2__20191216T215645_20191216T220909_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200315T065755_20200315T071241_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191030T135252_20191030T135600_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200219T081800_20200219T083303_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200110T203717_20200110T204612_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200409T053748_20200409T054151_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200413T053254_20200413T053659_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200208T191154_20200208T192117_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200319T065300_20200319T070802_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200304T175209_20200304T180102_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191128T122800_20191128T123212_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191009T150801_20191009T151142_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191121T231659_20191121T232817_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200215T082253_20200215T083741_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200121T094259_20200121T095800_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191005T151255_20191005T151621_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200427T150701_20200427T151544_E001.nc', \n",
+ " 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191024T004201_20191024T005059_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200308T174708_20200308T175621_E001.nc', \n",
+ " 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200402T162707_20200402T163602_E001.nc']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "eaa73f90-84f0-4429-854a-784c95afde66",
+ "metadata": {},
+ "source": [
+ "Download the CryoSat-2 files into your hub instance by calling the download_files function you imported from the script."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "ecef7122-b3d5-4ca0-b207-7ad3c35ca34d",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "download_files(user_email, esa_files)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6e9d27c7-d9d9-40ae-aab6-34888aedfe8f",
+ "metadata": {},
+ "source": [
+ "Stashing the files in a data folder to keep our notebook directory less cluttered."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2f6c0779-77cc-4d60-a7a6-cb4a0ce1ea25",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "!mv CS_LTA__SIR*.nc data"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "aeb468e5-7a5a-456c-96cb-6808af3f5eaa",
+ "metadata": {},
+ "source": [
+ "#### Use `earthaccess` for querying and direct S3 access of ATL10\n",
+ "\n",
+ "First we authenticate using `earthaccess`"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2165876a-c5fa-4df9-b7f5-c82f01e7bdba",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "auth = earthaccess.login()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "fb7adbf2-1148-4075-8578-91e01fbeacfc",
+ "metadata": {},
+ "source": [
+ "Then we use a spatial filter to search for ATL10 granules that intersect our area of interest. This is the same area we used in our cs2eo query above."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "37139d13-43e4-4563-b436-2879c76677ec",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "results = earthaccess.search_data(\n",
+ " short_name = 'ATL10',\n",
+ " version = '006',\n",
+ " cloud_hosted = True,\n",
+ " bounding_box = (-17, 79, 12, 83),\n",
+ " temporal = ('2019-10-01','2020-04-30'),\n",
+ " count = 10\n",
+ ")\n",
+ "#note that with count=10 we're limiting the number of files we actually access for this tutorial."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "b883fc7f-1c26-471c-88c4-664fa59bed16",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "display(results[1])"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e58e1111-1b07-47ec-952b-036290346f4c",
+ "metadata": {},
+ "source": [
+ "We use earthaccess.open() to directly access the ATL10 files within their S3 bucket. earthaccess.open() creates a file-like object, which is required because AWS S3 uses object storage, and we need to create a virtual file system to work with the HDF5 library."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "a80212ab-ee66-4d06-8a4a-47169092159e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "%time\n",
+ "icesat2_files = earthaccess.open(results) \n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "d24f9d50-3f95-4cdf-b877-1ac06be4471b",
+ "metadata": {},
+ "source": [
+ "We can use xarray to examine the contents of our files (one group at a time)."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "1d1102e5-561e-41e8-81df-e4ff543d19c1",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds_is2 = xr.open_dataset(icesat2_files[1], group='gt2r/freeboard_segment')\n",
+ "ds_is2"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "c603f6a6-7dcb-4a6f-9130-0bab933ff3c6",
+ "metadata": {},
+ "source": [
+ "And we can use hvplot to plot one of the variables within that group."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "0503dfee-7d66-49fa-8482-ad2dc330ff52",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds_is2['beam_fb_height'].hvplot(kind='scatter', s=2)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6a89c207-028f-47df-b7d9-5723bd5e836f",
+ "metadata": {},
+ "source": [
+ "#### Open and plot downloaded CryoSat-2 data \n",
+ "\n",
+ "We need a list of the downloaded CryoSat-2 files."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "60c1704a-0b17-4d4f-a7a8-dac8fe5b9701",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "downloaded_files = os.listdir(path)\n",
+ "downloaded_files"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "2d280de0-5de7-4a0c-9986-c94ab6b6d05a",
+ "metadata": {},
+ "source": [
+ "We use xarray to access the contents of our netcdf file. In this case, we are not \"streaming\" data from an S3 bucket, but are accessing the data locally."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "8657ed36-d583-46b9-8ed2-9b1b57cb5786",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds_cs2 = xr.open_dataset(path + downloaded_files[0])\n",
+ "ds_cs2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "795b0bdd-0895-4de4-8008-b6f15fcf2b3e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds_cs2['radar_freeboard_20_ku'].hvplot(kind='scatter', s=2)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "40a8164d-f5c1-4049-aefe-aeb08295092b",
+ "metadata": {},
+ "source": [
+ "#### Plot ICESat-2 and CryoSat-2 Freeboards on same map"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "ad5f7f2c-2ff8-47cc-8ea8-337f9183c20e",
+ "metadata": {},
+ "source": [
+ "Here we're plotting one file from each data set to save time."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "11476d13-0f1f-44f7-a364-4a949554d8c2",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "projection = ccrs.Stereographic(central_latitude=90.,\n",
+ " central_longitude=-45.,\n",
+ " true_scale_latitude=70.)\n",
+ "extent = [-2500000.000, 2500000., -2500000., 2500000.000]\n",
+ "\n",
+ "\n",
+ "fig = plt.figure(figsize=(10,10))\n",
+ "ax = fig.add_subplot(projection=projection)\n",
+ "ax.set_extent(extent, projection)\n",
+ "ax.add_feature(cfeature.OCEAN)\n",
+ "ax.add_feature(cfeature.LAND)\n",
+ "ax.gridlines(draw_labels=True)\n",
+ "\n",
+ "vmin = 0.\n",
+ "vmax = 1.\n",
+ "\n",
+ "# Plot Cryosat freeboard\n",
+ "cs2_img = ax.scatter(ds_cs2.lon_poca_20_ku, ds_cs2.lat_poca_20_ku, 5,\n",
+ " c=ds_cs2.radar_freeboard_20_ku, \n",
+ " vmin=vmin, vmax=vmax, # Set max and min values for plotting\n",
+ " cmap='Reds', # shading='auto' to avoid warning\n",
+ " transform=ccrs.PlateCarree()) # coords are lat,lon but map if NPS \n",
+ "\n",
+ "# Plot IS2 freeboard \n",
+ "is2_img = ax.scatter(ds_is2.longitude, ds_is2.latitude, 5,\n",
+ " c=ds_is2.beam_fb_height, \n",
+ " vmin=vmin, vmax=vmax, \n",
+ " cmap='Purples', \n",
+ " transform=ccrs.PlateCarree())\n",
+ "\n",
+ "# Add colorbars\n",
+ "fig.colorbar(cs2_img, label='Cryosat-2 Radar Freeboard (m)')\n",
+ "fig.colorbar(is2_img, label='ICESat-2 Lidar Freeboard (m)')"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "392bb8fd-8ec6-4fd4-b6db-dc0386094c7f",
+ "metadata": {},
+ "source": [
+ "Here we're plotting several ICESat-2 and CryoSat-2 files at a time. This takes a few minutes to render."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "4815c13b-88c3-4d10-850e-02cf766d1844",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "%%time\n",
+ "\n",
+ "# NSIDC WGS84 Polar Stereographic \n",
+ "projection = ccrs.Stereographic(central_latitude=90.,\n",
+ " central_longitude=-45.,\n",
+ " true_scale_latitude=70.)\n",
+ "extent = [-2500000.000, 2500000., -2500000., 2500000.000]\n",
+ "\n",
+ "fig = plt.figure(figsize=(10,10))\n",
+ "ax = fig.add_subplot(projection=projection)\n",
+ "ax.set_extent(extent, projection)\n",
+ "# ax.coastlines()\n",
+ "ax.add_feature(cfeature.OCEAN)\n",
+ "ax.add_feature(cfeature.LAND)\n",
+ "\n",
+ "ax.gridlines(draw_labels=True)\n",
+ "\n",
+ "vmin = 0.\n",
+ "vmax = 1.\n",
+ "\n",
+ "# Plot CryoSat-2 freeboards\n",
+ "for fp in downloaded_files:\n",
+ " ds = xr.open_dataset(path + fp)\n",
+ " cs2= plt.scatter(ds.lon_poca_20_ku, ds.lat_poca_20_ku, 5,\n",
+ " c=ds.radar_freeboard_20_ku, cmap=\"Reds\",\n",
+ " vmin=vmin, vmax=vmax,\n",
+ " transform=ccrs.PlateCarree())\n",
+ "\n",
+ "# Plot ICESat-2 freeboards\n",
+ "for fp in icesat2_files:\n",
+ " ds = xr.open_dataset(fp, group='gt2r/freeboard_segment')\n",
+ " is2 = plt.scatter(ds.longitude, ds.latitude, 5,\n",
+ " c=ds.beam_fb_height, cmap=\"Purples\",\n",
+ " vmin=vmin, vmax=vmax,\n",
+ " transform=ccrs.PlateCarree())\n",
+ " \n",
+ "fig.colorbar(cs2, label=\"CryoSat-2 Radar Freeboard (m)\")\n",
+ "fig.colorbar(is2, label=\"ICESat-2 Lidar Freeboard (m)\")\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "a4681430-14b4-4a97-9a0a-9f3f587dfb8a",
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "Python 3 (ipykernel)",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.10.14"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/notebooks/ICESat-2_Cloud_Access/README.md b/notebooks/ICESat-2_Cloud_Access/README.md
index c95904b..b77b3fd 100644
--- a/notebooks/ICESat-2_Cloud_Access/README.md
+++ b/notebooks/ICESat-2_Cloud_Access/README.md
@@ -1,24 +1,50 @@
# ICESat-2 Cloud Access
## Summary
-This notebook demonstrates searching for cloud-hosted ICESat-2 data and directly accessing Land Ice Height (ATL06) granules from an Amazon Compute Cloud (EC2) instance using the `earthaccess` package. NASA data "in the cloud" are stored in Amazon Web Services (AWS) Simple Storage Service (S3) Buckets. **Direct Access** is an efficient way to work with data stored in an S3 Bucket when you are working in the cloud. Cloud-hosted granules can be opened and loaded into memory without the need to download them first. This allows you take advantage of the scalability and power of cloud computing.
+We provide several notebooks showcasing how to search and access ICESat-2 from the NASA Earthdata Cloud. NASA data "in the cloud" are stored in Amazon Web Services (AWS) Simple Storage Service (S3) Buckets. **Direct Access** is an efficient way to work with data stored in an S3 Bucket when you are working in the cloud. Cloud-hosted granules can be opened and loaded into memory without the need to download them first. This allows you take advantage of the scalability and power of cloud computing.
+
+### [Accessing and working with ICESat-2 data in the cloud](./ATL06-direct-access_rendered.ipynb)
+This notebook demonstrates searching for cloud-hosted ICESat-2 data and directly accessing Land Ice Height (ATL06) granules from an Amazon Compute Cloud (EC2) instance using the `earthaccess` package.
+
+#### Key Learning Objectives
+1. Use `earthaccess` to search for ICESat-2 data using spatial and temporal filters and explore search results;
+2. Open data granules using direct access to the ICESat-2 S3 bucket;
+3. Load a HDF5 group into an `xarray.Dataset`;
+4. Visualize the land ice heights using `hvplot`.
+
+### [Plotting ICESat-2 and CryoSat-2 Freeboards](./ICESat2-CryoSat2.ipynb)
+This notebook demonstrates plotting ICESat-2 and CryoSat-2 data in the same map from within an AWS ec2 instance. ICESat-2 data are accessed via "direct S3 access" using `earthaccess`. CryoSat-2 data are downloaded to our cloud instance from their ftp storage lcoation and accessed locally.
+
+#### Key Learning Objectives
+1. use `earthaccess` to search for ICESat-2 ATL10 data using a spatial filter
+2. open cloud-hosted files using direct access to the ICESat-2 S3 bucket;
+3. use cs2eo script to download files into your hub instance
+3. load an HDF5 group into an `xarray.Dataset`;
+4. visualize freeboards using `hvplot`.
+5. map the locations of ICESat-2 and CryoSat-2 freeboards using `cartopy`
+
+### [Processing Large-scale Time Series of ICESat-2 Sea Ice Height in the Cloud](./ATL10-h5coro_rendered.ipynb)
+This notebook utilizes several libraries to performantly search, access, read, and grid ATL10 data over the Ross Sea, Antarctica including `earthaccess`, `h5coro`, and `geopandas`. The notebook provides further guidance on how to scale this analysis to the entire continent, running the same workflow from a script that can be run from your laptop using [Coiled](https://www.coiled.io/).
+
+#### Key Learning Objectives
+1. Use earthaccess to authenticate with Earthdata Login, search for ICESat-2 data using spatial and temporal filters, and directly access files in the cloud.
+2. Open data granules using h5coro to efficiently read HDF5 data from the NSIDC DAAC S3 bucket.
+3. Load data into a geopandas.DataFrame containing geodetic coordinates, ancillary variables, and date/time converted from ATLAS Epoch.
+4. Grid track data to EASE-Grid v2 6.25 km projected grid using drop-in-the-bucket resampling.
+5. Calculate mean statistics and assign aggregated data to grid cells.
+6. Visualize aggregated sea ice height data on a map.
## Set up
-To run the notebook provided in this folder in the Amazon Web Services (AWS) cloud, there are a couple of options:
-* An EC2 instance already set up with the necessary software installed to run a Jupyter notebook, and the environment set up using the provided environment.yml file. **Note:** If you are running this notebook on your own AWS EC2 instance using the environment set up using the environment.yml file in the NSIDC-Data-Tutorials/notebooks/ICESat-2_Cloud_Access/environment folder, you may need to run the following command before running the notebook to ensure the notebook executes properly:
+To run the notebooks provided in this folder in the Amazon Web Services (AWS) cloud, there are a couple of options:
+* An EC2 instance already set up with the necessary software installed to run a Jupyter notebook, and the environment set up using the provided environment.yml file. **Note:** If you are running these notebooks on your own AWS EC2 instance using the environment set up using the environment.yml file in the NSIDC-Data-Tutorials/notebooks/ICESat-2_Cloud_Access/environment folder, you may need to run the following command before running the notebook to ensure the notebook executes properly:
`jupyter nbextension enable --py widgetsnbextension`
You do NOT need to do this if you are using the environment set up using the environment.yml file from the NSIDC-Data-Tutorials/binder folder.
-* Alternatively, if you have access to one, it can be run in a managed cloud-based Jupyter hub. Just make sure all the necessary libraries are installed (`earthaccess`,`xarray`, and `hvplot`).
+* Alternatively, if you have access to one, it can be run in a managed cloud-based Jupyter hub. Just make sure all the necessary libraries are installed (e.g. `earthaccess`,`xarray`,`hvplot`, etc.).
-For further details on the prerequisites, see the 'Prerequisites' section in the notebook.
+For further details on the prerequisites, see the 'Prerequisites' section in each notebook.
-## Key Learning Objectives
-1. Use `earthaccess` to search for ICESat-2 data using spatial and temporal filters and explore search results;
-2. Open data granules using direct access to the ICESat-2 S3 bucket;
-3. Load a HDF5 group into an `xarray.Dataset`;
-4. Visualize the land ice heights using `hvplot`.
diff --git a/notebooks/ICESat-2_Cloud_Access/SIR_SAR_L2_E_download_script.py b/notebooks/ICESat-2_Cloud_Access/SIR_SAR_L2_E_download_script.py
new file mode 100644
index 0000000..a4d564a
--- /dev/null
+++ b/notebooks/ICESat-2_Cloud_Access/SIR_SAR_L2_E_download_script.py
@@ -0,0 +1,63 @@
+import os
+import platform
+from ftplib import FTP
+import sys
+
+
+download_file_obj = None
+read_byte_count = None
+total_byte_count = None
+
+
+def get_padded_count(count, max_count):
+ return str(count).zfill(len(str(max_count)))
+
+
+def file_byte_handler(data):
+ global download_file_obj, read_byte_count, total_byte_count
+ download_file_obj.write(data)
+ read_byte_count = read_byte_count + len(data)
+ progress_bar(read_byte_count, total_byte_count)
+
+
+def progress_bar(progress, total, prefix="", size=60, file=sys.stdout):
+ if total != 0:
+ x = int(size * progress / total)
+ x_percent = int(100 * progress / total)
+ file.write(f" {prefix} [{'='*x}{' '*(size-x)}] {x_percent} % \r")
+ file.flush()
+
+
+def download_files(user_email, esa_files):
+ global download_file_obj, read_byte_count, total_byte_count
+ print("About to connect to ESA science server")
+ with FTP("science-pds.cryosat.esa.int") as ftp:
+ try:
+ ftp.login("anonymous", user_email)
+ print("Downloading {} files".format(len(esa_files)))
+
+ for i, filename in enumerate(esa_files):
+ padded_count = get_padded_count(i + 1, len(esa_files))
+ print("{}/{}. Downloading file {}".format(padded_count, len(esa_files), os.path.basename(filename)))
+
+ with open(os.path.basename(filename), 'wb') as download_file:
+ download_file_obj = download_file
+ total_byte_count = ftp.size(filename)
+ read_byte_count = 0
+ ftp.retrbinary('RETR ' + filename, file_byte_handler, 1024)
+ print("\n")
+ finally:
+ print("Exiting FTP.")
+ ftp.quit()
+
+
+if __name__ == '__main__':
+
+ esa_files = ['SIR_SAR_L2/2019/12/CS_LTA__SIR_SAR_2__20191227T110305_20191227T111751_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200329T163208_20200329T164044_E001.nc', 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200114T203033_20200114T204440_E001.nc', 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191103T134759_20191103T135125_E001.nc', 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200204T191657_20200204T192558_E001.nc', 'SIR_SAR_L2/2019/12/CS_LTA__SIR_SAR_2__20191216T215645_20191216T220909_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200315T065755_20200315T071241_E001.nc', 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191030T135252_20191030T135600_E001.nc', 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200219T081800_20200219T083303_E001.nc', 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200110T203717_20200110T204612_E001.nc', 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200409T053748_20200409T054151_E001.nc', 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200413T053254_20200413T053659_E001.nc', 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200208T191154_20200208T192117_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200319T065300_20200319T070802_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200304T175209_20200304T180102_E001.nc', 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191128T122800_20191128T123212_E001.nc', 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191009T150801_20191009T151142_E001.nc', 'SIR_SAR_L2/2019/11/CS_LTA__SIR_SAR_2__20191121T231659_20191121T232817_E001.nc', 'SIR_SAR_L2/2020/02/CS_LTA__SIR_SAR_2__20200215T082253_20200215T083741_E001.nc', 'SIR_SAR_L2/2020/01/CS_LTA__SIR_SAR_2__20200121T094259_20200121T095800_E001.nc', 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191005T151255_20191005T151621_E001.nc', 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200427T150701_20200427T151544_E001.nc', 'SIR_SAR_L2/2019/10/CS_LTA__SIR_SAR_2__20191024T004201_20191024T005059_E001.nc', 'SIR_SAR_L2/2020/03/CS_LTA__SIR_SAR_2__20200308T174708_20200308T175621_E001.nc', 'SIR_SAR_L2/2020/04/CS_LTA__SIR_SAR_2__20200402T162707_20200402T163602_E001.nc']
+
+ if int(platform.python_version_tuple()[0]) < 3:
+ exit("Your Python version is {}. Please use version 3.0 or higher.".format(platform.python_version()))
+
+ email = input("Please enter your e-mail: ")
+
+ download_files(email, esa_files)
diff --git a/notebooks/ICESat-2_Cloud_Access/img/ATL10_CS2_L2_SAR_query_med.png b/notebooks/ICESat-2_Cloud_Access/img/ATL10_CS2_L2_SAR_query_med.png
new file mode 100644
index 0000000..1579b12
Binary files /dev/null and b/notebooks/ICESat-2_Cloud_Access/img/ATL10_CS2_L2_SAR_query_med.png differ
diff --git a/notebooks/ICESat-2_Cloud_Access/img/icesat2-cryosat2.png b/notebooks/ICESat-2_Cloud_Access/img/icesat2-cryosat2.png
new file mode 100644
index 0000000..b2ca77d
Binary files /dev/null and b/notebooks/ICESat-2_Cloud_Access/img/icesat2-cryosat2.png differ
 \n",
- " \n",
+ "\n",
+ "\n",
"# **Accessing and working with ICESat-2 data in the cloud**\n",
- " \n",
- "
\n",
- " \n",
+ "\n",
+ "\n",
"# **Accessing and working with ICESat-2 data in the cloud**\n",
- " \n",
- " \n",
- " \n",
+ "\n",
+ "\n",
"# **Accessing and working with ICESat-2 data in the cloud**\n",
- " \n",
- "
\n",
- " \n",
+ "\n",
+ "\n",
"# **Accessing and working with ICESat-2 data in the cloud**\n",
- " \n",
- "
 \n",
- "
\n",
- "