-
Beta Was this translation helpful? Give feedback.
Answered by
rtimms
Oct 13, 2022
Replies: 2 comments 1 reply
-
|
Here is an example import pybamm
import numpy as np
import matplotlib.pyplot as plt
pybamm.set_logging_level("INFO")
# load model
options = {
"current collector": "potential pair",
"dimensionality": 2, # 2D current collectors
"thermal": "x-lumped", # thermal model (ignores through-cell variation)
}
model = pybamm.lithium_ion.SPM(options)
# parameters can be updated here
param = model.default_parameter_values
# set mesh points
var = pybamm.standard_spatial_vars
var_pts = {
var.x_n: 10, # negative electrode
var.x_s: 10, # separator
var.x_p: 10, # positive electrode
var.r_n: 10, # negative particle
var.r_p: 10, # positive particle
var.y: 10, # current collector y-direction
var.z: 10, # current collector z-direction
}
# solver
solver = pybamm.CasadiSolver(atol=1e-6, rtol=1e-3, root_tol=1e-3, mode="fast")
# simulation
simulation = pybamm.Simulation(
model, parameter_values=param, var_pts=var_pts, solver=solver
)
# solve
t_eval = np.linspace(0, 3600, 100) # time in seconds
simulation.solve(t_eval=t_eval)
solution = simulation.solution
# plotting ---------------------------------------------------------------------
# post-process variables
phi_s_cn = solution["Negative current collector potential [V]"]
phi_s_cp = solution["Positive current collector potential [V]"]
I = solution["Current collector current density [A.m-2]"]
T = solution["X-averaged cell temperature [K]"]
# mesh for plotting
L_y = param.evaluate(model.param.L_y)
L_z = param.evaluate(model.param.L_z)
y_plot = np.linspace(0, L_y, 21)
z_plot = np.linspace(0, L_z, 21)
# define plotting function
def plot(t):
fig, ax = plt.subplots(figsize=(15, 8))
plt.tight_layout()
plt.subplots_adjust(left=-0.1)
# negative current collector potential
plt.subplot(221)
phi_s_cn_plot = plt.pcolormesh(
y_plot, z_plot, phi_s_cn(y=y_plot, z=z_plot, t=t).T, shading="gouraud"
)
plt.axis([0, L_z, 0, L_z])
plt.xlabel(r"$y$ [m]")
plt.ylabel(r"$z$ [m]")
plt.title(r"$\phi_{s,cn}$ [V]")
plt.set_cmap("cividis")
plt.colorbar(phi_s_cn_plot)
# positive current collector potential
plt.subplot(222)
phi_s_cp_plot = plt.pcolormesh(
y_plot, z_plot, phi_s_cp(y=y_plot, z=z_plot, t=t).T, shading="gouraud"
)
plt.axis([0, L_z, 0, L_z])
plt.xlabel(r"$y$ [m]")
plt.ylabel(r"$z$ [m]")
plt.title(r"$\phi_{s,cp}$ [V]")
plt.set_cmap("viridis")
plt.colorbar(phi_s_cp_plot)
# through-cell current
plt.subplot(223)
I_plot = plt.pcolormesh(
y_plot, z_plot, I(y=y_plot, z=z_plot, t=t).T, shading="gouraud"
)
plt.axis([0, L_z, 0, L_z])
plt.xlabel(r"$y$ [m]")
plt.ylabel(r"$z$ [m]")
plt.title(r"$I$ [A.m-2]")
plt.set_cmap("plasma")
plt.colorbar(I_plot)
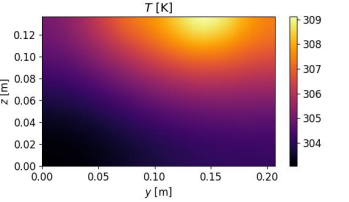
# temperature
plt.subplot(224)
T_plot = plt.pcolormesh(
y_plot, z_plot, T(y=y_plot, z=z_plot, t=t).T, shading="gouraud"
)
plt.axis([0, L_z, 0, L_z])
plt.xlabel(r"$y$ [m]")
plt.ylabel(r"$z$ [m]")
plt.title(r"$T$ [K]")
plt.set_cmap("inferno")
plt.colorbar(T_plot)
plt.subplots_adjust(
top=0.92, bottom=0.15, left=0.10, right=0.9, hspace=0.5, wspace=0.5
)
plt.show()
# call plot with time in seconds
plot(1000)you can evaluate the post-processed variables at any y, z, t. |
Beta Was this translation helpful? Give feedback.
1 reply
Answer selected by
rtimms
-
|
Beta Was this translation helpful? Give feedback.
0 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Here is an example