Replies: 3 comments 3 replies
-
|
Hi @tlsw231, sorry about your weird simulation bug 😢 Can you please post a MCVE, ideally using fake data? I'm surprised that |
Beta Was this translation helpful? Give feedback.
-
|
you should be able to write a preprocess function that fixes the time coordinate. I'd imagine it to be something like
I'm convinced there's a more straight-forward way, but my initial attempt would look like this: In [61]: time_coord = [0, 0.25, 0.5, 0.75, 1.0, 3221249121, 1.5, 1.75, 2, 0.009223241, 2.5, 2.75]
...: ds = xr.Dataset(coords={"time": time_coord})
...: ds
Out[61]:
<xarray.Dataset>
Dimensions: (time: 12)
Coordinates:
* time (time) float64 0.0 0.25 0.5 0.75 1.0 ... 1.75 2.0 0.009223 2.5 2.75
Data variables:
*empty*
In [62]: time = ds.time.reset_index("time", drop=True)
...: padded = (
...: ds.time.reset_index("time", drop=True)
...: .pad({"time": (1, 1)}, mode="constant", constant_values=float("nan"))
...: .interpolate_na(dim="time", method="linear", fill_value="extrapolate")
...: )
...: mask = (
...: padded.rolling({"time": 3}, center=True)
...: .construct(window_dim="window")
...: .isel(time=slice(1, -1))
...: .diff(dim="window")
...: .pipe(lambda da: (da == 0.25).any(dim="window"))
...: )
...: ds.assign_coords(time=time.where(mask).interpolate_na(dim="time", method="linear")).time
Out[62]:
<xarray.DataArray 'time' (time: 12)>
array([0. , 0.25, 0.5 , 0.75, 1. , 1.25, 1.5 , 1.75, 2. , 2.25, 2.5 , 2.75])
Coordinates:
* time (time) float64 0.0 0.25 0.5 0.75 1.0 ... 1.75 2.0 2.25 2.5 2.75 |
Beta Was this translation helpful? Give feedback.
-
|
But xr.open_mfdataset, even with combine='nested', sorts the timestamps which ends up changing the original relative location of the garbage coordinate values (not desirable).
I don't think this is intended behaviour. If this can be reproduced with a
MVCE, I can treat it as a bug report.
|
Beta Was this translation helpful? Give feedback.
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
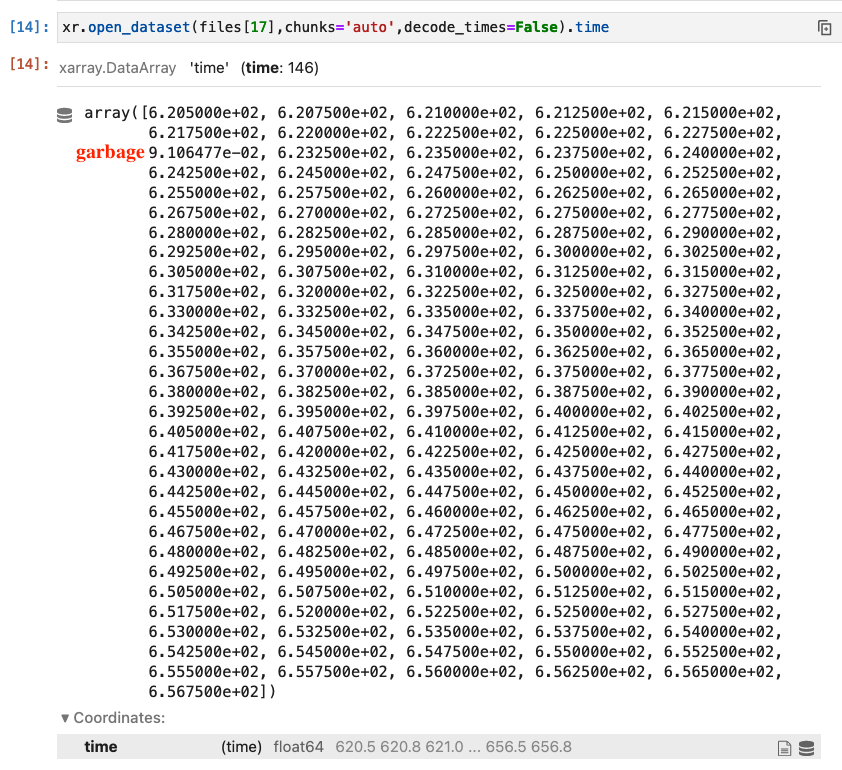
I am facing a slightly unusual data salvaging problem. I have a climate dataset with perfectly good data (temperature, humidity, etc.) but due to some weird Intel-related bug during the simulation, the values of the coordinates
timehave gotten messed up. Here is how they look when I run the NCO toolncdump -v timefrom the command line on one of the output files generated:0, 0.25, 0.5, 0.75, 1., 3221249121, 1.5, 1.75, 2, 0.009223241, 2.5, 2.75............where the
timecoordinate is measuring the time elapsed (in days) from some reference date. The fields are written out every 6 hours (= 0.25 days), except that every once in a while, the time coordinate value is complete garbage (due to that bug). In the example above, the garbage coordinate values should be 1.25 and 2.25, respectively.The way to get around this would be to find the problematic positions using
np.where(np.diff(ds.time) != 0.25)[0]and substitute the garbage values at those positions with the average value of the adjacent values.I tried reading in the dataset with
combine='nested'anddecode_cf=Falsewhich prevents the conversion of thetimecoordinate toDatetimeobjects. So far, so good. But to execute the simple solution above, I don't wantxarrayto sort using thetimecoordinate values. But that is exactly what happens and after I read in the datasetds.timelooks like this:0, 0.009223241, 0.25, 0.5, 0.75, 1., 1.5, 1.75, 2, 2.5, 2.75............3221249121,....Without knowing the correct position of the garbage values, I now cannot just use the average value of the adjacent values.
Is there any way to read in the dataset such that xarray leaves the
timecoordinated unsorted?[PS: 1. It is probably possible to handle this purely with NCO tools but I am still exploring ways to do this within xarray.
2. Regenerating the data is out of the question as these are high-resolution climate runs spanning more than a century, i.e., computationally expensive.]
Beta Was this translation helpful? Give feedback.
All reactions