Replies: 1 comment 5 replies
-
|
Thanks for reporting. Do you have a small example to reproduce? My guess here is that your transformations on the |
Beta Was this translation helpful? Give feedback.
5 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
-
Hi!
So I have graphs in the PyG format -
PyG graph DataBatch(x=[554, 11], edge_index=[2, 1320], edge_attr=[1320, 4], y=[31, 19], pos=[554, 3], idx=[31], name=[31], z=[554], batch=[554], ptr=[32])
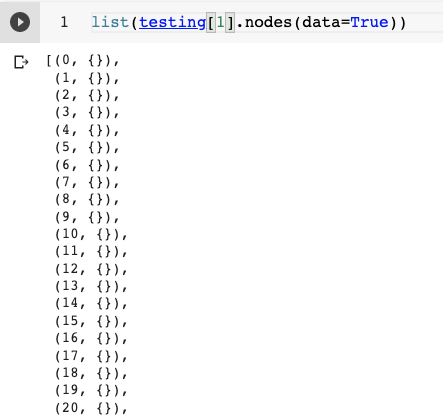
I convert them to networkx graphs as follows -
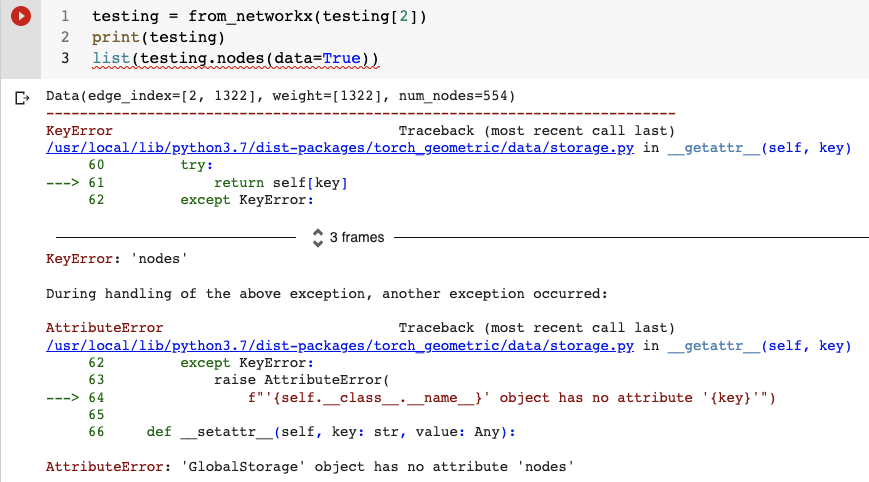
to_networkx(graph , node_attrs=['x'], graph_attrs=['y']))On these NetworkX graphs I have to perform some manipulations, like adding edge/deletions based on some criteria and save them back to PyG graphs. But on doing this, I am not able to get the original labels back, basically I would want to be able to work on the modified graph using some GNN.
After manipulating the graph if I convert them back to PyG graphs this is what I get -
Data(edge_index=[2, 1322], weight=[1322], num_nodes=554)The original information is lost. Is there any way to get the original labels back? Any insights are appreciated.
Thank you!
Beta Was this translation helpful? Give feedback.
All reactions