Replies: 1 comment 3 replies
-
|
We just added support for temporal-based link sampling within If you are batching multiple graphs, you can also leverage the seed_node_time = …
mask = edge_time < seed_node_time[batch[edge_index[0]]
edge_index = edge_index[:, mask] |
Beta Was this translation helpful? Give feedback.
3 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
-
Hi team,
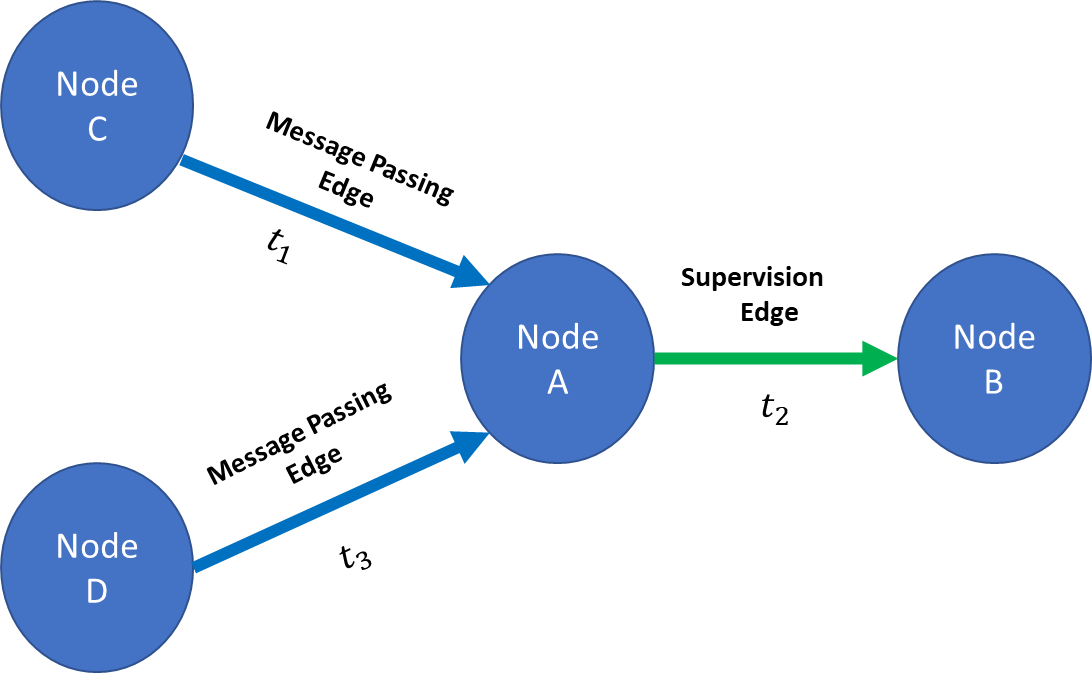
I'm working on an edge classification problem with temporal constraints.
For example, given a supervision-edge from time t1 I would like to filter out all neighboring edges that came from t>t1 ("the future of t1") to prevent data leakage.
I'm having trouble performing this type of filtering on the batch level (meaning, filtering each supervision-edge's neighboring message-passing edges using the supervision-edges' timestamps) because the message-passing edges defined by
edge_indexare not stored separately for each sample in the batch. (edge_indexhas a[2, num_edges]dimensions).I tried the following two approaches:
edge_indexby the single supervision edge's timestamp.Worked extremely slow.
For each batch:
The loss is aggregated for each time window and the optimizer step happens at the end of each batch.
Works faster.
If anybody has a more elegant solution I would be happy to hear : )
Beta Was this translation helpful? Give feedback.
All reactions