-
Notifications
You must be signed in to change notification settings - Fork 0
Home
Welcome to the iEEGLAB wiki!
- via the extension manager
[to add after release is made]
- via Github download
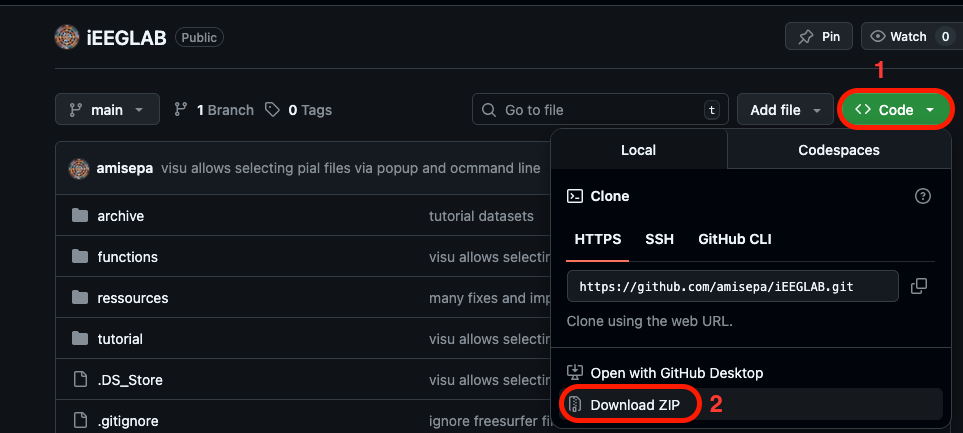
- via Git clone
Navigate to your local eeglab folder (where you installed it) > plugins folder and clone the repo there with:
git clone https://github.com/amisepa/iEEGLAB.git
Dataset description: This sEEG dataset is subject 2 from this BIDS dataset: https://nemar.org/dataexplorer/detail?dataset_id=ds004696 The dataset in brief, patients were resting in the hospital bed, while single pulse stimulation was performed. The stimulation had a duration of 200 μs, was biphasic and had an amplitude of 6mA. See publication here for more detail on the dataset and the Cortico-cortical evoked potentials (CCEP) methods: https://pmc.ncbi.nlm.nih.gov/articles/PMC10538586/ For tutorial purposes, we:
- removed the first 369 s of data before the experiment started,
- adjusted events timestamps accordingly
- kept only 19 min of experiment data
- downsampled the .mefd dataset from 2048 to 128 Hz
- exported it as an EEGLAB .set dataset
We end up with 16 electrodes and 169 (single-pulse stimulation) events.
Open MATLAB and open EEGLAB by typing eeglab in your MATLAB command window:
eeglab
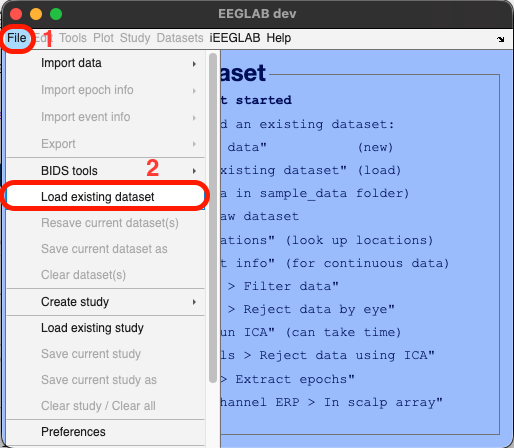
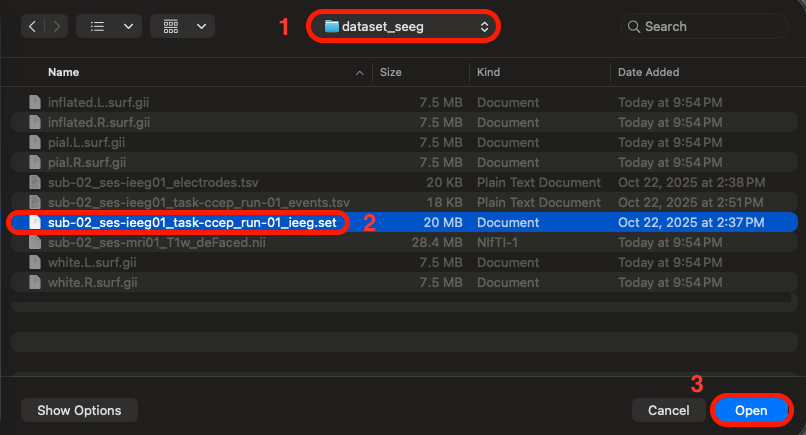
Navigate to File > Load existing dataset > navigate to eeglab > plugins > iEEGLAB > tutorial > dataset_seeg > select the file named "sub-02_ses-ieeg01_task-ccep_run-01_ieeg.set" > click open
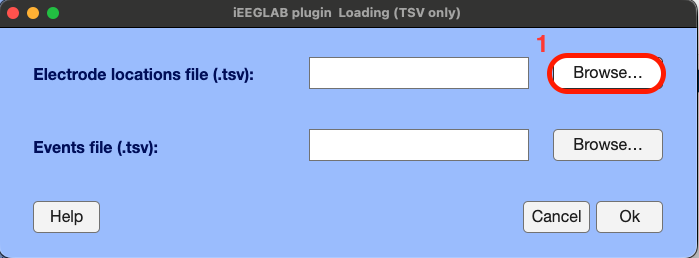
In EEGLAB, go to the iEEGLAB tab > Load electrode coordinates and events

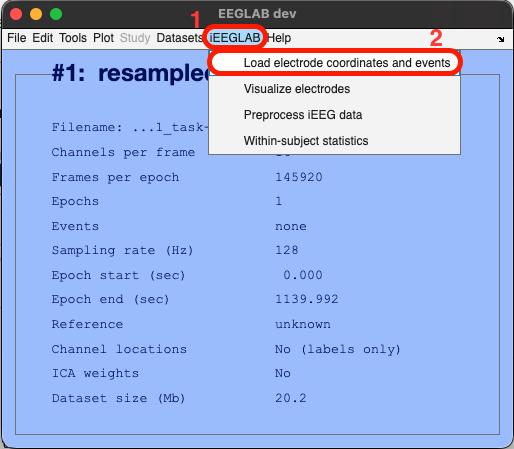
Loading electrodes coordinates is mandatory, whereas loading events is optional (if events are already contained by EEGLAB dataset depending on how the data were collected, or if the application is continuous like epilepsy or medical monitoring).
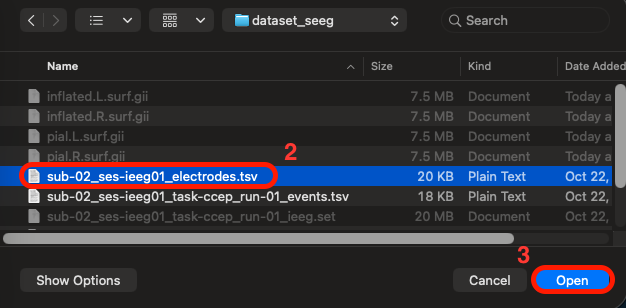
Click the "Browse" button and select the tsv file containing the sEEG electrodes spatial 3D (XYZ) coordinates, named "sub-02_ses-ieeg01_electrodes.tsv" > click "Open"
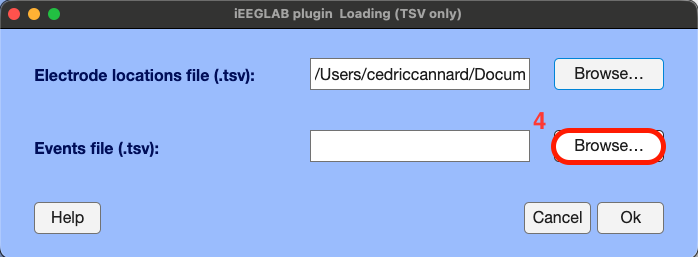
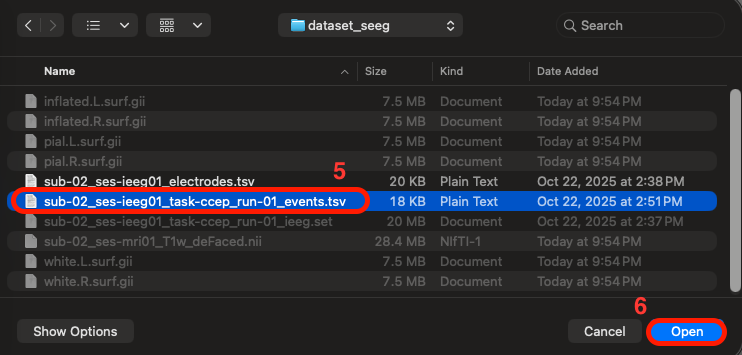
Same for the events, Click "Browse" > select the tsv file containing the event markers, named "sub-02_ses-ieeg01_task-ccep_run-01_events.tsv" > Click "Ok"
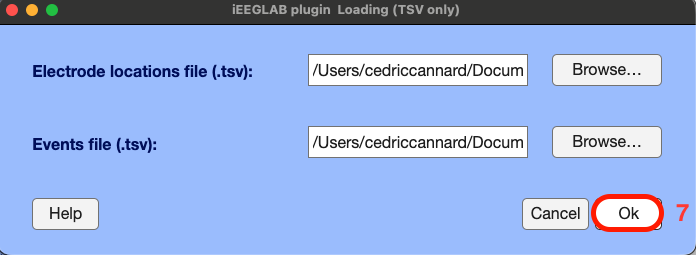
A new EEGLAB interface pops-up automatically to select:
- electrodes of interest (e.g., if you wish to only analyze a few specific electrodes)
- the event field of interest from the .tsv file (by default the code detects the stimulation site by electrode name, but if you wish you can switch to stimulation site by number, or whichever field you may have in your .tsv events file).
- the events of interest (if you wish to only analyze specific conditions/events/stimuli).
Here, we leave the default "All" to include all electrodes and events for subsequent analysis.
And we see the following outputs in MATLAB's command window:
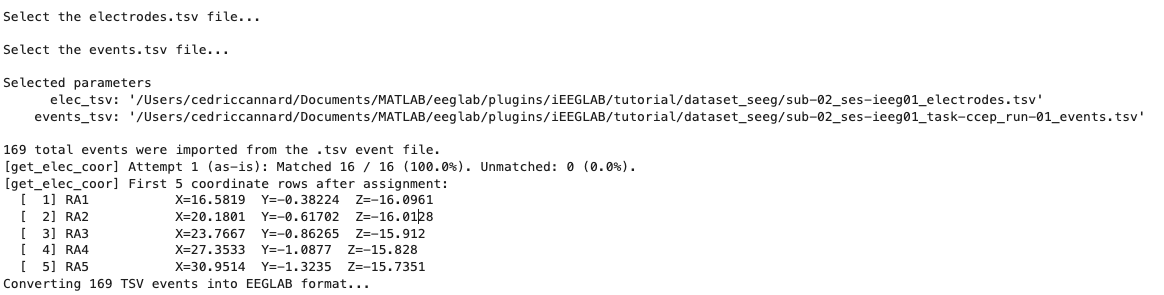
iEEGLAB automatically loaded the electrode XYZ coordinates and events from the two .tsv files, matched the electrode labels to ensure the spatial coordinates match the correct electrode (with 2 fallback methods for best success), runs checks, etc. Here we can see that 100% of electrodes were matched. Note that by default, to make these steps faster and easier, the iEEGLAB plugin will take you to the folder where you loaded the EEGLAB dataset. Also, the plugin supports .csv files, if you do not have these .tsv files. IMPORTANT: if your dataset already contains events (stored in EEG.event after loading the data into EEGLAB), you do not need to select a events.tsv file.
You can launch all these steps via one command line:
EEG = ieeglab_load(EEG);
The tutorial dataset comes with the necessary Pial Surface files (one for each hemisphere) generated by Freesurfer from the subject's T1 MRI, we just need to select them from the iEEGLAB interface. How to generate these surface files with Freesurfer is shown later (a process which can take several hours of computation).
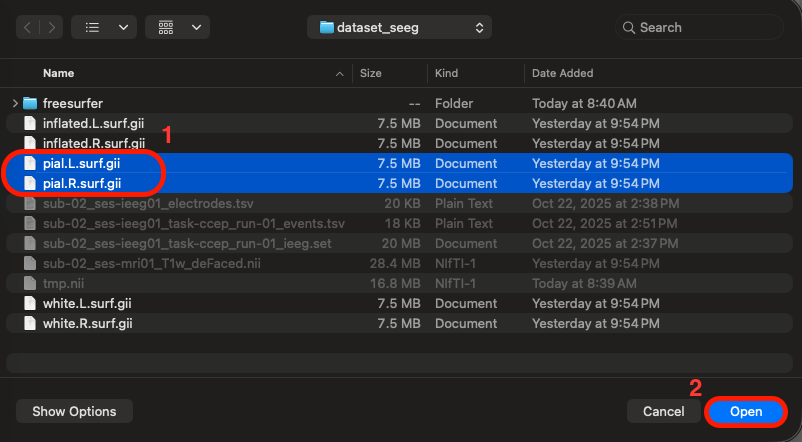
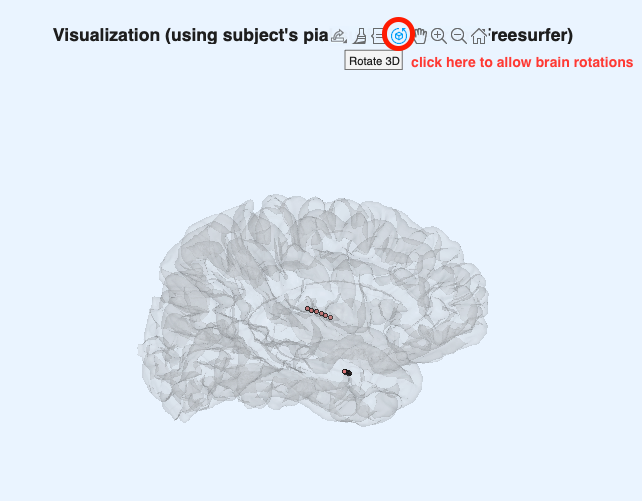
Go to the iEEGLAB tab in EEGLAB > Click on "Visualize electrodes" > Select the two pial.surf files (using the CTRL/COMMAND key to select both; note that "L" and "R" indicate left/right hemispheres, respectively) > Click Open > The glass brain figure appears, showing the 16 sEEG electrode sites located in the right hemisphere (remember this is a reduced dataset for tutorial purposes, but the full dataset has 256 electrodes spread out in both hemispheres. The iEEGLAB plugin only plots one hemisphere if all available electrodes are located in only one hemisphere).
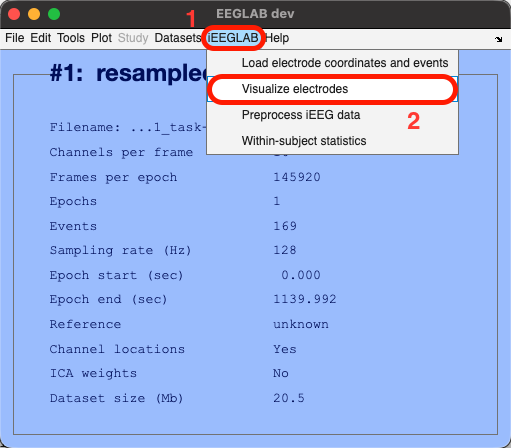
Via command line:
ieeglab_vis_elec(EEG);
- eCoG dataset

eCoG_glassbrain_pial.mp4
- sEEG dataset

sEEG_glassbrain_pial.mp4
If the participant's pial surface file is not available, it falls back to dipfit's standard_BEM (smooth) template. This is of course much less accurate as it does not account for individual differences in brain/head size, shape, etc.
- eCoG dataset
eCoG_glassbrain_nopial.mp4
- sEEG dataset

sEEG_glassbrain_nopial.mp4
- Visualize raw time series and events (sEEG CCEP dataset)

Usual EEGLAB command line:
pop_eegplot(EEG,1,1,1);