-
-
Notifications
You must be signed in to change notification settings - Fork 63
Python tight binding
sids implements an easy data object for creating tight-binding models.
The easiest way to describe the tight-binding model is showcase its usage.
The graphene structure is one of the easiest structures to describe and provide tight-binding parameters for.
First we create the geometry with nearest neighbour interactions
import sids
# Lattice constant
alat = 1.42
length = ( 1.5 ** 2 + 3. / 4. ) ** .5 * alat
C = sids.Atom(6,R=alat+0.1) # add small number for numerics
sc = sids.SuperCell([length,length,10,90,90,60],nsc=[3,3,1])
# Rotate supercell as the initial one is x = cartesian
sc = sc.rotate(-30,v=[0,0,1])
gr = sids.Geometry([[0,0,0],[1,0,0]], atoms=C, sc=sc)
which now results in a graphene unit cell with appropriate periodicity for nearest neighbour interactions.
We now proceed to create the tight-binding object
tb = sids.TightBinding(gr)
now we have a tight-binding object, but no hopping integrals or on-site energies have been set.
# Denote interactions ranges
# on-site , nearest neighbour
dR = (0.1 , alat + 0.1)
for ia in gr:
# Get interacting atoms
idx = gr.close(ia, dR=dR)
tb[ia,idx[0]] = ( 0. , 1.)
tb[ia,idx[1]] = (-2.7, 0.)
there are a few things going on here:
-
dRis an array of arbitrary length which can be understood as spherical radii for returning the atoms close to another atom or position.The array can be arbitrarily long and returns a list of equal length with each list entry contain all entries in the spherical shell defined by the previous radii and the current radii.
In the above code
idxreturned fromcloseis-
idx[0]all atoms with distance <= .1 of atomia -
idx[1]all atoms 0.1 < distance <= alat + 0.1 of atomia
-
-
tb[ia,idx[0]] = (0.,1.)sets the on-site energy to0and the overlap matrix to1. -
tb[ia,idx[1]] = (-2.7,0)sets the nearest neighbour hopping integral to-2.7and the overlap matrix to0, hence yielding an orthogonal tight-binding model.
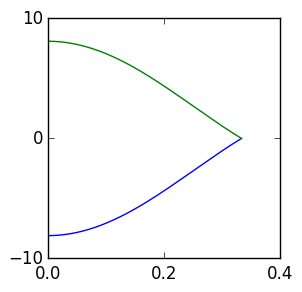
To finalise the bandstructure we calculate the bandstructure from
\Gamma to K (so not full band-structure).
nk = 200
kpts = np.linspace(0, 1./3, nk)
eigs = np.empty([nk,2],np.float64)
for ik, k in enumerate(kpts):
eigs[ik,:] = tb.eigh(k=[k,-k,0])
import matplotlib.pyplot as plt
plt.plot(kpts,eigs[:,0])
plt.plot(kpts,eigs[:,1])
plt.show()
Which results in this band-structure
Press this source link to get the complete source.