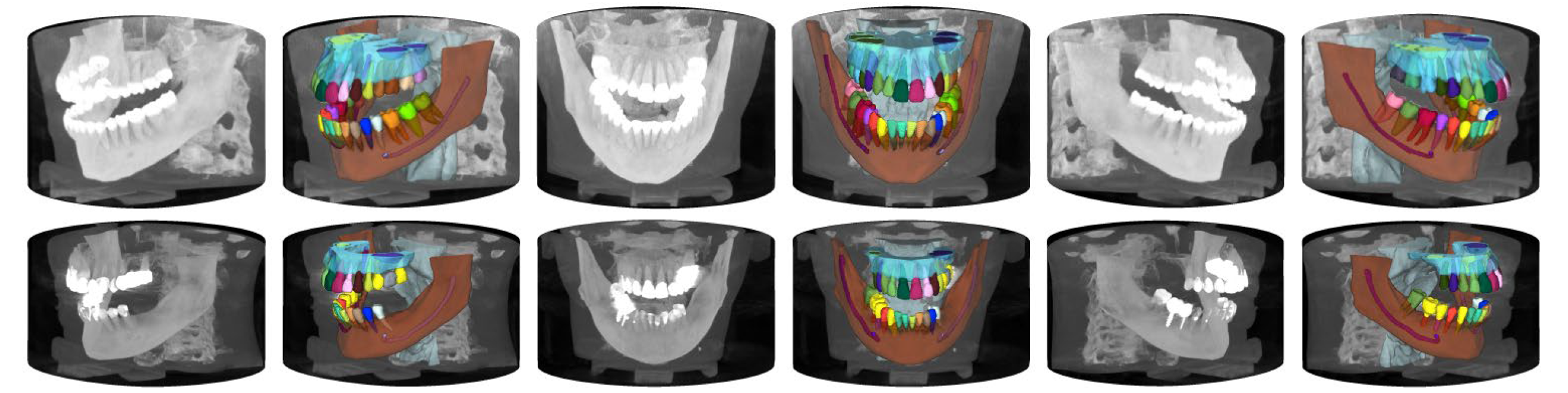
ToothFairy2-CVPR is the official benchmark repository for the ToothFairy2 dataset, a large-scale, publicly available collection of CBCT scans with voxel-level 3D annotations for 42 maxillofacial structures, including teeth, jawbones, sinuses, and alveolar canals. This benchmark evaluates state-of-the-art segmentation methods—ranging from CNNs to transformers and Mamba-based models on maxillofacial anatomical regions, using the ToothFairy2 dataset for training and test.
All benchmarked methods are implemented within the nnU-Net framework, with targeted architectures implemented by us in the benchmark_networks/nnunetv2/nets folder and their corresponding trainer classes in the benchmark_networks/nnunetv2/training/nnUNetTrainer directory.
To train models with nnU‑Net based architecture, organize your dataset according to the nnU‑Net dataset format, as specified in the official documentation here. Specifically, you need to:
- Create dataset folder
Create the nnUNet_raw, nnUNet_preprocessed and nnUNet_results folders, and set the corresponding environment variables:
export nnUNet_raw_data_base="<path to nnUNet_raw>"
export nnUNet_preprocessed="<path to nnUNet_preprocessed>"
export RESULTS_FOLDER="<path for trained models>"- File structure
Place your data in:
nnUNet_raw/DatasetXXX_ToothFairy2/
├── imagesTr/
├── labelsTr/
└── [optional] imagesTs/
The naming of the single dataset cases must be:
- Training images:
caseID_0000.nii.gz - Corresponding labels:
caseID.nii.gz(samecaseID)
- Generate
dataset.json
In the dataset root folder, include adataset.jsonfile like. You can auto-generate it using:
python -m nnunetv2.dataset_conversion generate_dataset_json -o nnUNet_raw/DatasetXXX_ToothFairy2 - Plan and preprocess To verify the dataset format, compute dataset fingerprints, prepare preprocessed data for training, and generate plans files, run the following commands (you can find more details here):
nnUNetv2_plan_and_preprocess -d XXX --verify_dataset_integrityActually, we already provide our plans file in the nnUNetplans_files folder.
To train the different models on the ToothFairy2 dataset (replace XXX with your dataset ID), use the following commands (more details here):
# Standard nnUNet (3D full resolution)
nnUNetv2_train XXX 3d_fullres 0 -tr nnUNetTrainer -p nnUNetPlans
# ResEncL nnUNet version (3D full resolution)
nUNetv2_train XXX 3d_fullres 0 -tr nnUNetTrainer -p nnUNetResEncUNetL
# nnFormer (3D full resolution)
nnUNetv2_train XXX 3d_fullres 0 -tr nnUNetTrainerUMmabaBot -p nnUNetPlans
# SwinUMamba (2D)
nnUNetv2_train XXX 2d 0 -tr nnUNetTrainerSwinUMambaD -p nnUNetPlans
# VMamba (2D)
nnUNetv2_train XXX 2d 0 -tr nnUNetTrainerVmamba -p nnUNetPlans
# UMamba (3D full resolution)
nnUNetv2_train XXX 3d_fullres 0 -tr nnUNetTrainerUMmabaBot -p nnUNetPlans
# Training without left/right mirroring (substitute the {nnUNetTrainerName} with one of the trainer names above, and correspondent 2d/3d_fullres configuration)
srun nnUNetv2_train XXX 2d/3d_fullres 0 -tr {nnUNetTrainerName}_onlyMirror01 -p nnUNetPlans