-
Notifications
You must be signed in to change notification settings - Fork 0
Update documentation (minor) #83
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Changes from 10 commits
5378bfa
a5ab28e
f5ce2b7
5f0e666
6cf6bb3
f9575cc
06fb961
3af33ee
798c48c
64939d7
582d974
File filter
Filter by extension
Conversations
Jump to
Diff view
Diff view
There are no files selected for viewing
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -36,7 +36,11 @@ An overview of the VueGen workflow is shown in the figure below: | |
|
|
||
| 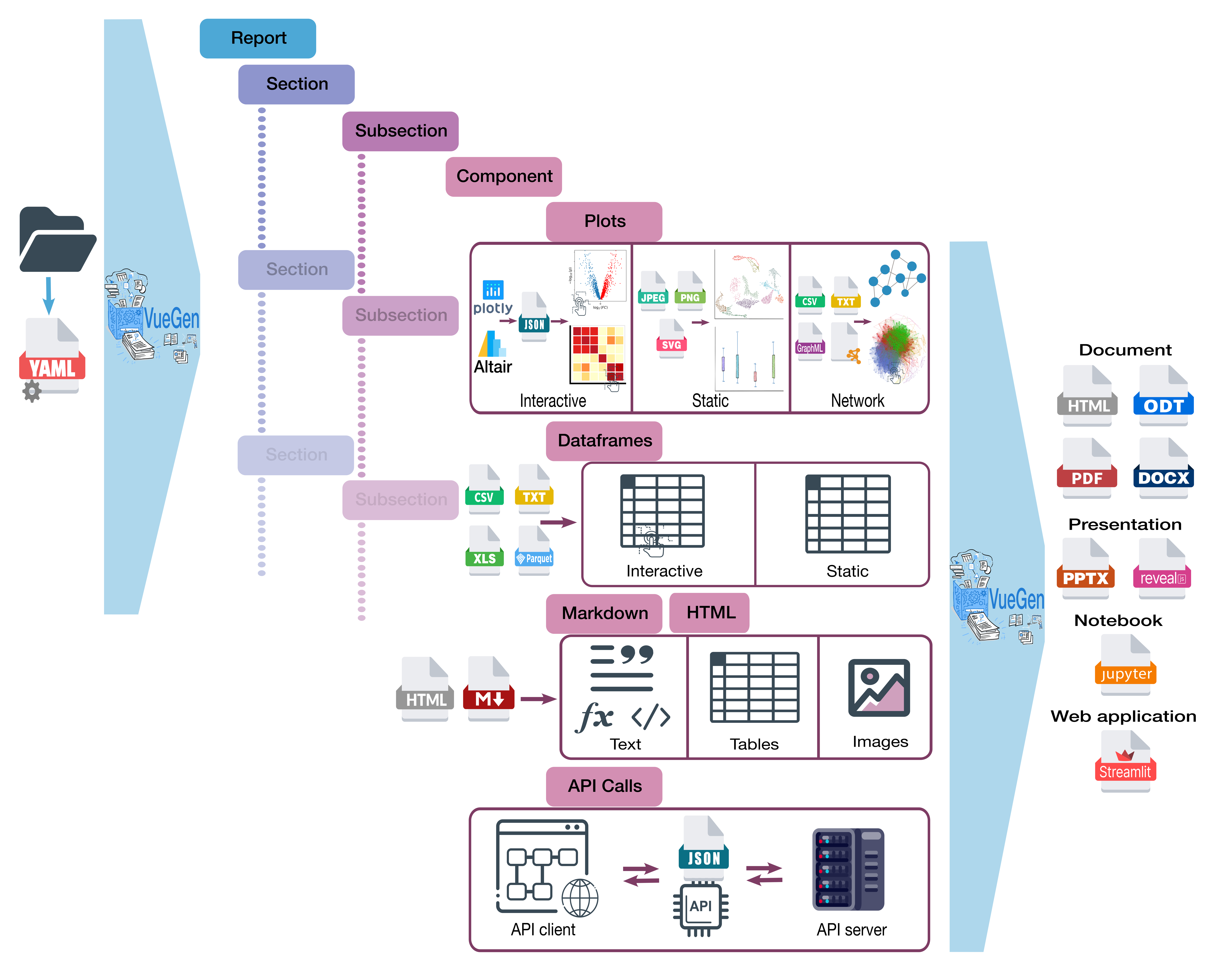 | ||
|
|
||
| Also, the class diagram for the project is presented below to illustrate the architecture and relationships between classes: | ||
| We created a schema diagram to illustrates the structure of the configuration file and the relationships between its elements: | ||
|
|
||
| 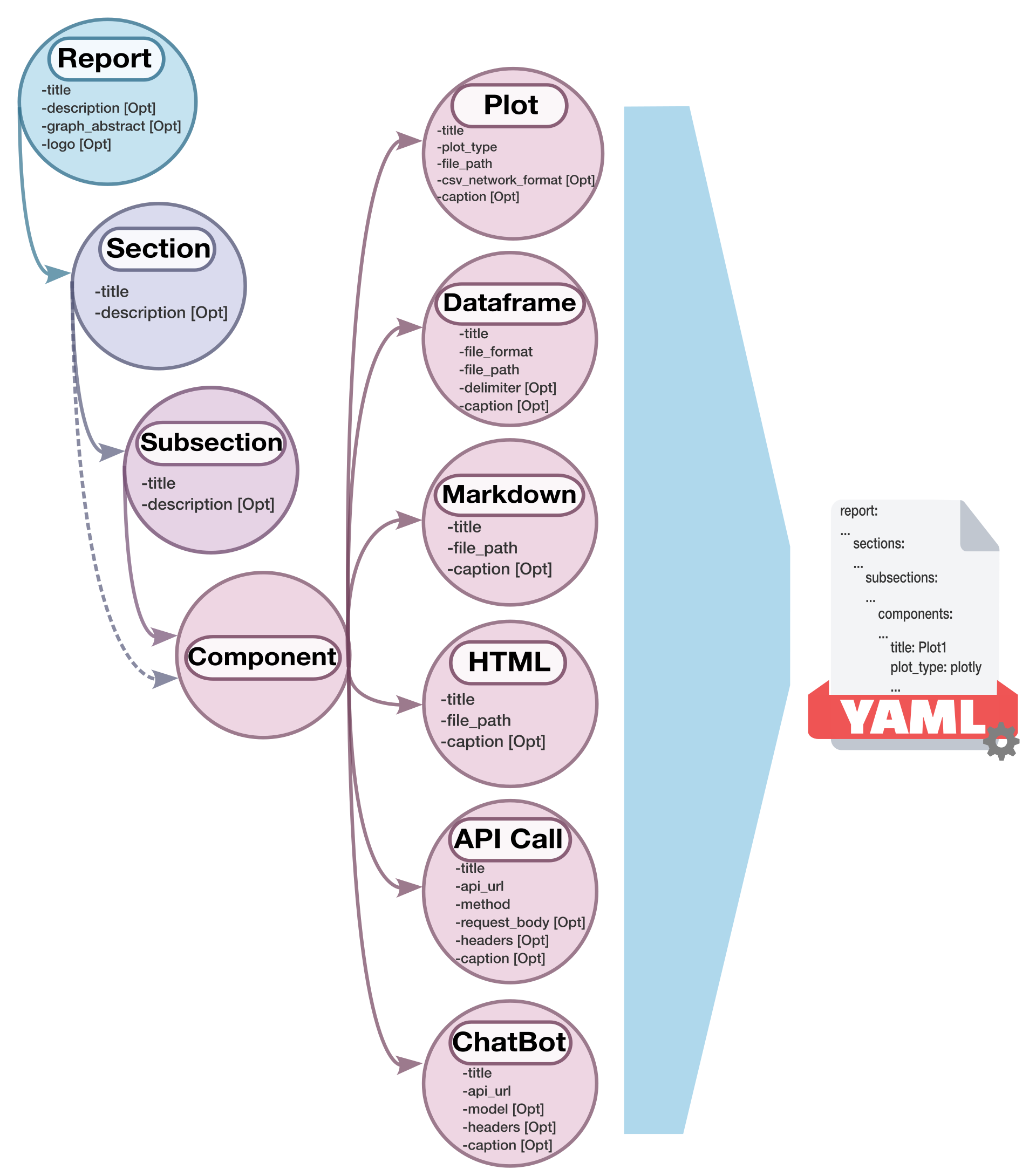 | ||
|
|
||
| Also, the class diagram for the project's current version is presented below to show the architecture and relationships between classes: | ||
|
|
||
| 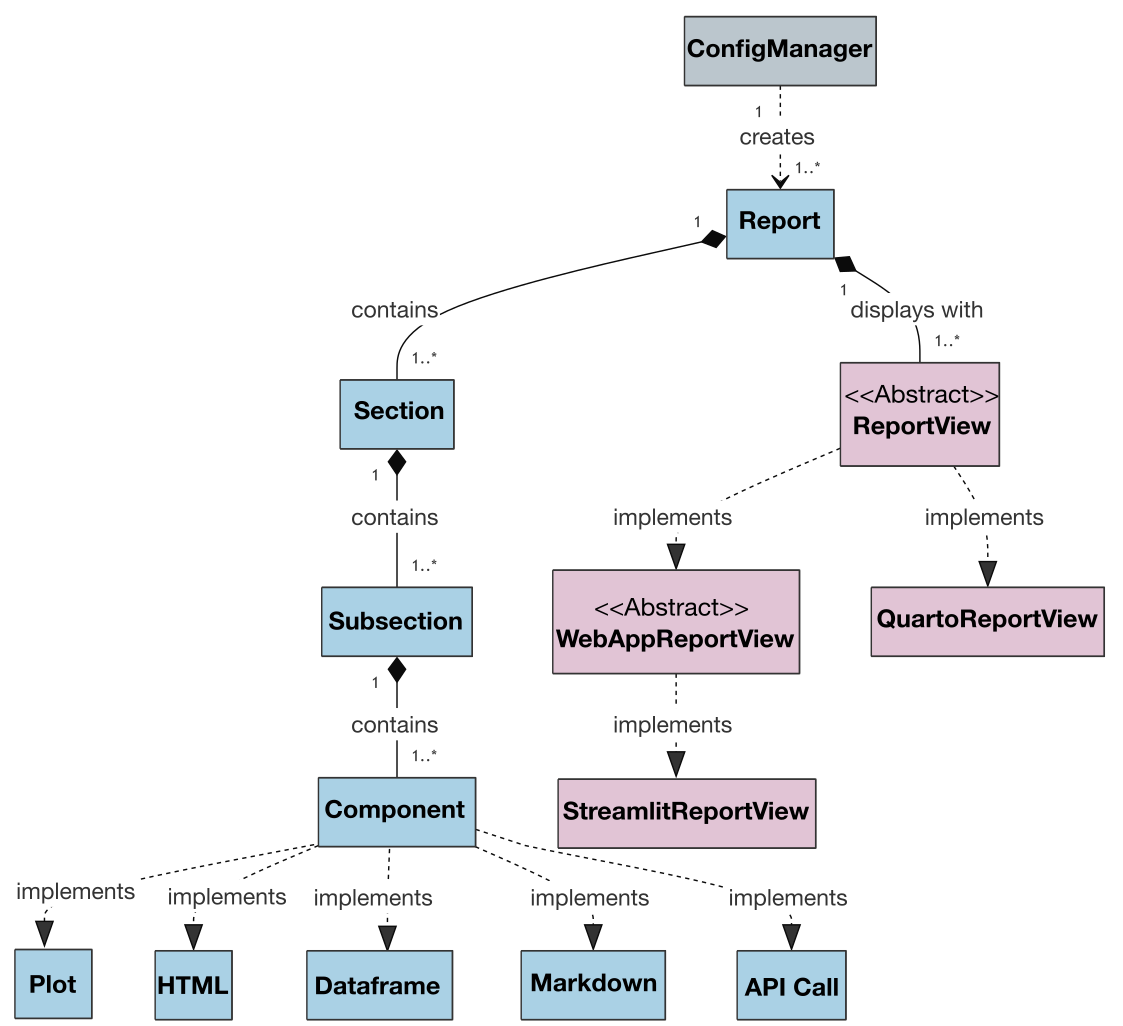 | ||
|
|
||
|
|
@@ -59,9 +63,11 @@ pip install vuegen | |
|
|
||
| You can also install the package for development by cloning this repository and running the following command: | ||
|
|
||
| > [!WARNING] | ||
| > We assume you are in the root directory of the cloned repository when running this command. Otherwise, you need to specify the path to the `vuegen` directory. | ||
|
|
||
| ```bash | ||
| pip install -e path/to/vuegen # specify location | ||
| pip install -e . # in case your pwd is in the vuegen directory | ||
| pip install -e . | ||
| ``` | ||
|
|
||
| ### Conda | ||
|
|
@@ -98,12 +104,19 @@ If you prefer not to install VueGen on your system, a pre-configured Docker cont | |
|
|
||
| ### Nextflow and nf-core | ||
|
|
||
| VueGen is also available as a [nf-core][nfcore] module, customised for compatibility with the [Nextflow][nextflow] environment. This module is designed to automate report generation from outputs produced by other modules, subworkflows, or pipelines. You can read the offical documentation for the nf-core module [here](nf-vuegen-nf-core). Also, the source code and detailed documentation are available in the [nf-VueGen repository][nf-vuegen]. | ||
| VueGen is also available as a [nf-core][nfcore] module, customised for compatibility with the [Nextflow][nextflow] environment. This module is designed to automate report generation from outputs produced by other modules, subworkflows, or pipelines. Asumming that you have `nextflow` and `nf-core` installed, you can use the following command to install the nf-core module: | ||
|
|
||
| ```bash | ||
| nf-core modules install vuegen | ||
| ``` | ||
|
|
||
| > [!NOTE] | ||
| > You can read the offical documentation for the nf-core module [here][nf-vuegen-nf-core]. Also, the source code and additional details are available in the [nf-VueGen repository][nf-vuegen]. | ||
|
|
||
| ## Execution | ||
|
|
||
| > [!IMPORTANT] | ||
| > Here we use the `Earth_microbiome_vuegen_demo_notebook` [directory][emp-dir] and the `Earth_microbiome_vuegen_demo_notebook.yaml` [configuration file][emp-config] as examples, which are available in the `docs/example_data` and `docs/example_config_files` folders, respectively. Make sure to clone this reposiotry to access these contents, or use your own directory and configuration file. | ||
| > Here we use the `Earth_microbiome_vuegen_demo_notebook` [directory][emp-dir] and the `Earth_microbiome_vuegen_demo_notebook.yaml` [configuration file][emp-config] as examples, which are available in the `docs/example_data` and `docs/example_config_files` folders, respectively. Make sure to clone the VueGen's GitHub reposiotry to access these contents, or use your own directory and configuration file. | ||
|
|
||
| Run VueGen using a directory with the following command: | ||
|
|
||
|
|
@@ -116,18 +129,20 @@ vuegen --directory docs/example_data/Earth_microbiome_vuegen_demo_notebook --rep | |
|
|
||
| ### Folder structure | ||
|
|
||
| Your input directory must follow a **nested folder structure**, where first-level folders are treated as **sections** and second-level folders as **subsections**, containing the components (plots, tables, networks, Markdown text, and HTML files). | ||
| Your input directory should follow a **nested folder structure**, where first-level folders are treated as **sections** and second-level folders as **subsections**, containing the components (plots, tables, networks, Markdown text, and HTML files). If the component files are in the first-level folders, an `overview` subsection will be created automatically. | ||
|
|
||
| Here is an example layout: | ||
|
|
||
| ``` | ||
| report_folder/ | ||
| ├── section1/ | ||
| │ ├── table1.tsv | ||
| │ └── subsection1/ | ||
| │ ├── table.csv | ||
| │ ├── table2.csv | ||
| │ ├── image1.png | ||
| │ └── chart.json | ||
| ├── section2/ | ||
| │ ├── image2.jpg | ||
| │ ├── subsection1/ | ||
| │ │ ├── summary_table.xls | ||
| │ │ └── network_plot.graphml | ||
|
|
@@ -136,9 +151,6 @@ report_folder/ | |
| │ └── summary.md | ||
| ``` | ||
|
|
||
| > [!WARNING] | ||
| > VueGen currently requires each section to contain at least one subsection folder. Defining only sections (with no subsections) or using deeper nesting levels (i.e., sub-subsections) will result in errors. In upcoming releases, we plan to support more flexible directory structures. | ||
|
|
||
| The titles for sections, subsections, and components are extracted from the corresponding folder and file names, and afterward, users can add descriptions, captions, and other details to the configuration file. Component types are inferred from the file extensions and names. | ||
| The order of sections, subsections, and components can be defined using numerical suffixes in folder and file names. | ||
|
|
||
|
|
@@ -172,16 +184,57 @@ docker run --rm \ | |
| quay.io/dtu_biosustain_dsp/vuegen:v0.3.2-docker --directory /home/appuser/Earth_microbiome_vuegen_demo_notebook --report_type streamlit | ||
| ``` | ||
|
|
||
| ### Running VueGen with Nextflow and nf-core | ||
|
|
||
| To run VueGen as a nf-core module, you should create a Nextflow pipeline and include the VueGen module in your workflow. Here is a `main.nf` example: | ||
|
|
||
| ```groovy | ||
| #!/usr/bin/env nextflow | ||
| include { VUEGEN } from './modules/nf-core/vuegen/' | ||
|
|
||
| workflow { | ||
| // Create a channel for the report type | ||
| report_type_ch = Channel.value(params.report_type) | ||
|
|
||
| // Handle configuration file and directory inputs | ||
| if (params.config) { | ||
| file_ch = Channel.fromPath(params.config) | ||
| input_type_ch = Channel.value('config') | ||
| output_ch = VUEGEN(input_type_ch, file_ch, report_type_ch) | ||
|
|
||
| } else if (params.directory) { | ||
| dir_ch = Channel.fromPath(params.directory, type: 'dir', followLinks: true) | ||
| input_type_ch = Channel.value('directory') | ||
| output_ch = VUEGEN(input_type_ch, dir_ch, report_type_ch) | ||
|
|
||
| } | ||
| } | ||
| ``` | ||
|
|
||
| You can run the pipeline with the following command: | ||
|
|
||
| ```bash | ||
| nextflow run main.nf --directory docs/example_data/Basic_example_vuegen_demo_notebook --report_type html | ||
| ``` | ||
|
|
||
| > [!NOTE] | ||
| > You can read the offical documentation for the nf-core module [here][nf-vuegen-nf-core]. Also, the source code and additional details are available in the [nf-VueGen repository][nf-vuegen]. | ||
|
|
||
| ## GUI | ||
|
|
||
| We have a simple GUI for VueGen that can be run locally or through a standalone executable. | ||
|
|
||
| ### Local GUI | ||
|
|
||
| We have a simple GUI for VueGen that can be run locally or through a standalone executable. | ||
| For now you will need to have a copy of this repository. | ||
| To use the **local GUI**, you should clone this repository and install the required dependencies. You can do this by running the following command in the root directory of the cloned repository: | ||
|
|
||
| ```bash | ||
| pip install '.[gui]' | ||
| cd gui | ||
| ``` | ||
|
|
||
| Then, you should move to the `gui` folder and execute the `app.py` Python file: | ||
|
|
||
| ```bash | ||
| python app.py | ||
| ``` | ||
|
|
||
|
|
@@ -190,53 +243,66 @@ python app.py | |
| The **bundle GUI** with the VueGen package is available under the | ||
| [latest releases](https://github.com/Multiomics-Analytics-Group/vuegen/releases/latest). | ||
| You will need to unzip the file and run `vuegen_gui` in the unpacked main folder. | ||
| Most dependencies are included into the bundle using PyInstaller. | ||
| Most dependencies are included into the bundle using [PyInstaller][pyinstaller]. | ||
|
Collaborator
Author
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. Did you want to set a link?
Collaborator
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. I just defined all links in the bottom, so we do not have them in the main md. |
||
|
|
||
| Streamlit works out of the box as a purely Python based package. For `html` creation you will have to | ||
| have a Python 3.12 installation with the `jupyter` package installed as `quarto` needs to start | ||
| Streamlit works out of the box as a purely Python based package. For the rest of report types you | ||
| will have to have a **Python 3.12** installation with the `jupyter` package installed, as `quarto` needs to start | ||
| a kernel for execution. This is also true if you install `quarto` globally on your machine. | ||
|
|
||
| We recommend using miniforge to install Python and the conda package manager: | ||
|
|
||
| - [conda-forge.org/download/](https://conda-forge.org/download/) | ||
| > [!TIP] | ||
| > It is advisable to create a virtual environment to manage depenendencies and avoid conflicts with existing packages. | ||
| You can use the virtual environment manager of your choice, such as `poetry`, `conda`, or `pipenv`. | ||
| We recommend using [miniforge][conda-download] to install Python and the `conda` package manager. | ||
|
|
||
| We continous our example assuming you have installed the `miniforge` distribution for your | ||
| machine (MacOS with arm64/ apple silicon or x86_64/ intel or Windows x86_64). Also download | ||
| the [latest `vuegen_gui` bundle](https://github.com/Multiomics-Analytics-Group/vuegen/releases/latest) | ||
| We assume you have installed the `miniforge` distribution for your machine (MacOS with arm64/ apple silicon | ||
| or x86_64/ intel or Windows x86_64). Also, download the | ||
| [latest `vuegen_gui` bundle](https://github.com/Multiomics-Analytics-Group/vuegen/releases/latest) | ||
| from the releases page according to your operating system. | ||
|
|
||
| You can create a new conda environment with Python 3.12 and the `jupyter` and `vuegen` package: | ||
|
|
||
| ```bash | ||
| conda create -n vuegen_gui -c conda-forge python=3.12 jupyter vuegen | ||
| ``` | ||
|
|
||
| Then, activate the environment: | ||
|
|
||
| ```bash | ||
| conda create -n vuegen_gui -c conda-forge python=3.12 jupyter | ||
| # in case you have errors, install vuegen addtionally | ||
| conda activate vuegen_gui | ||
| pip install vuegen | ||
| # list all conda environments to find the location of the environment | ||
| conda info -e # find environment location | ||
| ``` | ||
|
|
||
| Find the vuegen_gui path for your local `user`. | ||
| > [!WARNING] | ||
| > If you have errors with the `vuegen` package, you can install it separately using pip, | ||
| as explained in the installation section. | ||
|
|
||
| On **MacOS** you need to add a `bin` to the path: | ||
| Now, you can list all conda environments to find the location of the `vuegen_gui` environment: | ||
|
|
||
| ```bash | ||
| /Users/user/miniforge3/envs/vuegen_gui/bin | ||
| conda info -e | ||
| ``` | ||
|
|
||
| On **Windows** you can use the path as displayed by `conda info -e`: | ||
| On **MacOS**, you need to add a `bin` to the path: | ||
|
|
||
| > Note: On Windows a base installation of miniforge with `jupyter` might work as well as | ||
| > the app can see your entire Path which is not the case on MacOS. | ||
| ```bash | ||
| /Users/user/miniforge3/envs/vuegen_gui/bin | ||
| ``` | ||
|
|
||
| On **Windows**, you can use the path as displayed by `conda info -e`: | ||
|
|
||
| ```bash | ||
| C:\Users\user\miniforge3\envs\vuegen_gui | ||
| ``` | ||
|
|
||
| > [!NOTE] | ||
| > On Windows a base installation of miniforge with `jupyter` might work because | ||
| the app can see your entire Path, which is not the case on MacOS. | ||
|
|
||
| More information regarding the app and builds can be found in the | ||
| [GUI README](https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/gui/README.md). | ||
| [GUI README][gui-readme]. | ||
|
||
|
|
||
| ## Case studies | ||
|
|
||
| VueGen’s functionality is demonstrated through two case studies: | ||
| VueGen’s functionality is demonstrated through various case studies: | ||
|
|
||
| ### 1. Predefined Directory | ||
|
|
||
|
|
@@ -376,6 +442,9 @@ We appreciate your feedback! If you have any comments, suggestions, or run into | |
| [emp]: https://earthmicrobiome.org/ | ||
| [emp-config]: https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/docs/example_config_files/Earth_microbiome_vuegen_demo_notebook_config | ||
| [emp-dir]: https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/docs/example_data/Earth_microbiome_vuegen_demo_notebook | ||
| [conda-download]: https://conda-forge.org/download/ | ||
| [gui-readme]: https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/gui/README.md | ||
| [pyinstaller]: https://pyinstaller.org/ | ||
| [st-cloud]: https://streamlit.io/cloud | ||
| [stlite_repo]: https://github.com/whitphx/stlite | ||
| [st-forum-exe]: https://discuss.streamlit.io/t/streamlit-deployment-as-an-executable-file-exe-for-windows-macos-and-android/6812 | ||
|
|
||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
this is at least not rendered on readthedocs: https://vuegen--83.org.readthedocs.build/en/83/sections_readme/execution.html#running-vuegen-with-nextflow-and-nf-core
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think this part is in the Installation section