-
Notifications
You must be signed in to change notification settings - Fork 6
Pseudomonas validation #57
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
Note: commit messages ccf38bb and earlier are from a merge conflict from #54 that I addressed but resulted in my ajlee master branch getting out of sync with greenelab master. So these commits are already merged in greenelab master but still showing in the history. These commits do not reflect the current changes made in this PR |
 ben-heil
left a comment
ben-heil
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looks good! Sorry for leaving so many comments, it's late (for me) so I'm more confused than usual
This PR performs a similar validation, previously performed on recount2 data, to pseudomonas data. Here we are comparing the ranking of genes generated by SOPHIE vs those from a manually curated dataset, GAPE.
The following changes were made:
0_prepare_reference_gene_file.ipynbnotebook that processes the curated ANOVA results to get gene rankings. These gene rankings will be what we compare our SOPHIE rankings against2_identify_generic_genes_pathways.ipynbnotebook to compare SOPHIE rankings vs the manually curated ones.0_subset_training_compendium.ipynbto create new training compendium and2_identify_generic_genes_pathways_pao1.ipynbto perform validation analysis on the new training compendium. The code in this notebook is nearly identical to2_identify_generic_genes_pathways.ipynband so doesn't need much review. There were some cutsom edits that needed to be made due to limitations in ponyo. Which I have created an issue for.The main result is here:
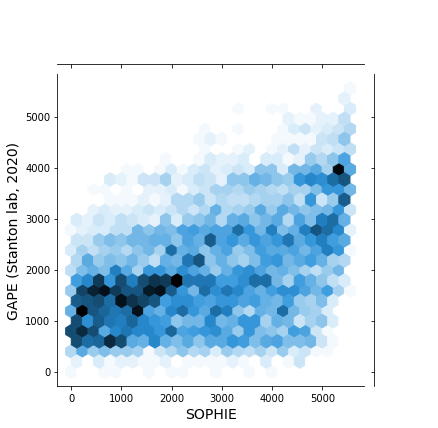
Using only PAO1 samples we get:
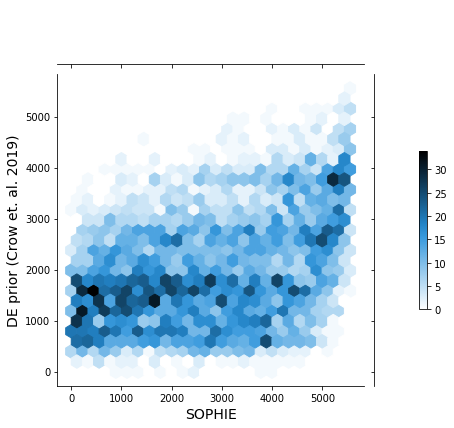
This inconsistency in genes found to be generic by SOPHIE and not by the manually curated set of experiments appears in this analysis using P. aeruginosa data and also human data. Some other hypotheses to test in the future include: