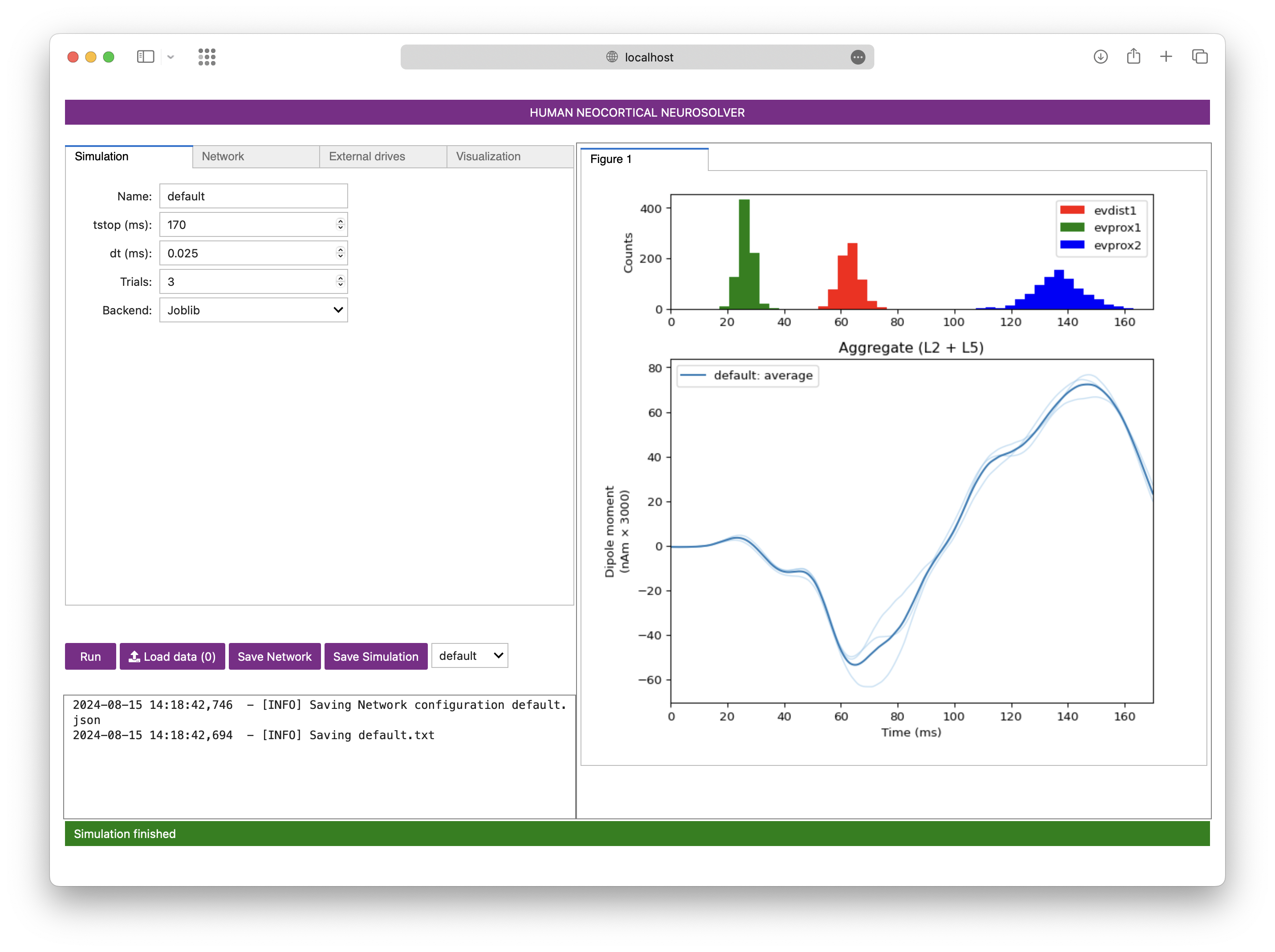
The Human Neocortical Neurosolver (HNN) is an open-source neural modeling tool designed to help researchers/clinicians interpret human brain imaging data. This repository, called HNN-core, houses the source code for HNN.
With only a few lines of code, HNN provides a convenient way to run simulations of an anatomically- and biophysically-detailed dynamical system model of human thalamocortical brain circuits. Given its modular, object-oriented design, HNN makes it easy to generate and evaluate hypotheses on the mechanistic origin of signals measured with magnetoencephalography (MEG), electroencephalography (EEG), or intracranial electrocorticography (ECoG). A unique feature of the HNN model is that it accounts for the biophysics generating the primary electric currents underlying such data. Simulation results are directly comparable to source-localized data (current dipoles in units of nano-Ampere-meters), enabling precise tuning of model parameters to match characteristics of recorded signals. Multimodal neurophysiology data such as local field potential (LFP), current-source density (CSD), and spiking dynamics can also be simulated simultaneously with current dipoles.
You can view HNN's frontpage here for an overview of all that HNN can do. For how to use HNN, we provide scientific documentation, tutorials, and examples aplenty on our HNN Textbook website. There, we describe the use of HNN in studying the circuit-level origin of some of the most commonly measured MEG/EEG and ECoG signals: event related potentials (ERPs) and low-frequency rhythms (alpha/beta/gamma).
The HNN API, written in Python and built on top of NEURON, is designed to be flexible and serve users with varying levels of coding expertise, while the HNN GUI is designed to be useful to researchers with no formal computational neural modeling or coding experience.
The terms HNN, HNN-core, and hnn-core are effectively equivalent, as they are all
different names for the same codebase. Historically, HNN-core was developed based on
the original, deprecated HNN repository, however that
repository is no longer supported or developed. It is kept online only for the sake
of scientific reproducibility.
Please consider supporting HNN development efforts by voluntarily providing your demographic information here! Note that any demographic information we collect is anonymized and aggregated for reporting on the grants that fund the continued development of HNN. All questions are voluntary.
You can try HNN in your browser for free, with no local installation required! At the top of our Installation Guide, you can find links that describe how to run HNN online in the cloud, either using Google CoLab notebooks or using the Neuroscience Gateway Portal.
To install HNN locally, see our Installation Guide located at the HNN Textbook
website. The easiest way to install hnn-core with the all its dependencies on Mac,
Linux, or Windows (using "Windows Subsystem for Linux"), is to first install the
Anaconda Python Distribution and then run
the following commands:
conda create -y -q -n hnn-core-env python=3.12
conda activate hnn-core-env
conda install hnn-core-all -c jonescompneurolab -c conda-forge
Our Anaconda packages currently only support Python 3.12. However, installing hnn-core
through pip currently supports Python 3.9 through 3.13, inclusively. Please see
our Installation Guide for detailed instructions on the various ways you can install
HNN.
Once you have installed hnn-core and the dependencies for the features you want, you
can find tutorials, examples, and scientific documentation at our HNN Textbook
website.
You can use the GitHub Issues tracker to report bugs. For user questions, installation help, and scientific discussions, please see our GitHub Discussions page.
Contributors are always welcome! Please read our Contributing Guide and make sure to abide by our Code of Conduct. Our governance structure can be found here.
If you use HNN-core in your work, please cite our publication in JOSS:
Jas et al., (2023). HNN-core: A Python software for cellular and circuit-level interpretation of human MEG/EEG. Journal of Open Source Software, 8(92), 5848, https://doi.org/10.21105/joss.05848
HNN-core development has been funded in part by the following United States of America federal government grants:
- NIH R01EB022889 Human Neocortical Neurosolver
- NIH 1U24NS129945 Dissemination of the Human Neocortical Neurosolver (HNN) software for circuit level interpretation of human MEG/EEG
- NSF IIS 2424101 Collaborative Research: CRCNS Research Proposal: Uncovering the mechanisms and meaning of brain rhythm frequency shifts during decision making