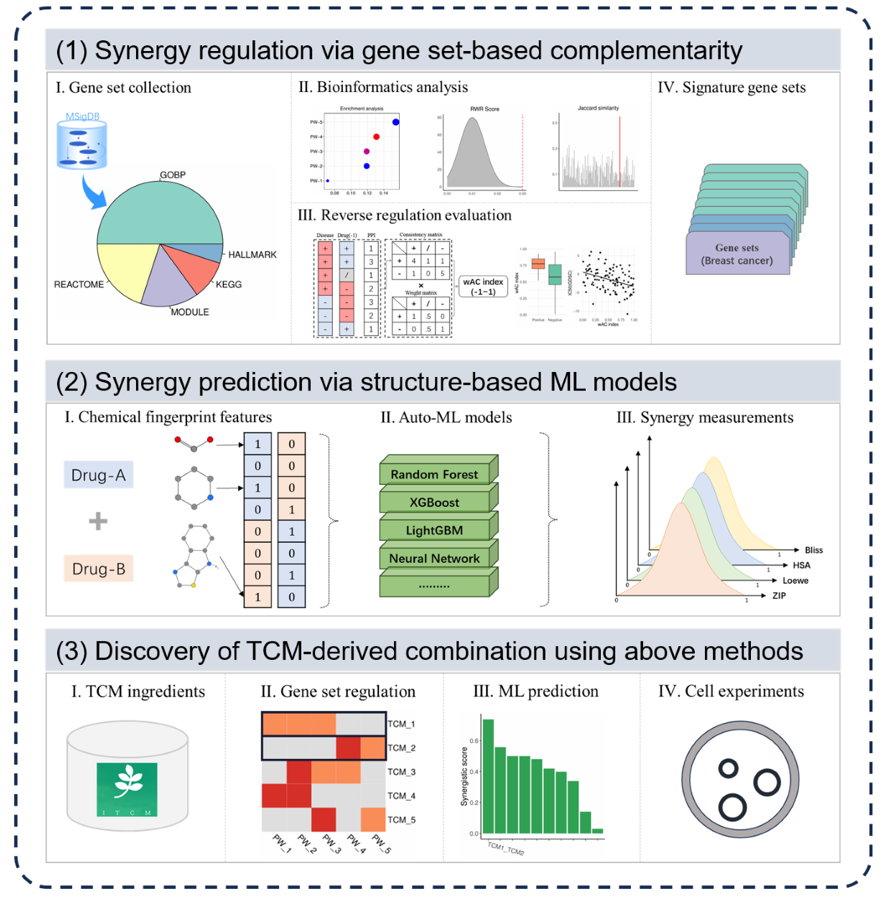
2.1 Propose computational algorithm based on Complementary signature regulation
- 1)TCGA-DEG identification and Over-representation enrichment analysis;
- 2)RAR proximity of Signature to breast cancer targets;
- 3)De-redundancy analysis between signatures;
- 4)Propose wAC index based on reversal regulation to recognize marker signature.
2.2 Propose computational algorithm based on Complementary signature regulation
- 1)data collection and cleaning;
- 2)AutoML model training;
- 3)Model comparison and selection.
2.3 Discover TCM compound combination
- 1)Signature regulation score of single TCM compound;
- 2)Signature regulation score of TCM compound combinations;
- 3)Overall ML prediction based on ZIP synergistic measurements;
- 4)Validation of synergy effect via a series of in vitro experiments
- Gene signatures: MSigDB, https://www.gsea-msigdb.org/gsea/msigdb/index.jsp
- Drug--Disease, Drug--Target, Target--Disease information: https://db.idrblab.net/ttd/full-data-download
- Protein–Protein Interactions: https://www.nature.com/articles/s41467-019-09186-x#Sec23, Supplementary Data 1
- Compound-perturbating signatures ;Metadata for signatures, genes, compounds: Expanded CMap LINCS Resource 2020 , https://clue.io/data/CMap2020#LINCS2020
- GDSC1-dataset, GDSC2-dataset: GDSC, https://www.cancerrxgene.org/downloads/bulk_download
- TCM High-throughput Data, TCM metadata: ITCM, http://itcm.biotcm.net/download.html
- Drug Combination Measurement: SYNERGxDB, https://www.synergxdb.ca/synergy_score?&dataset=2
- Breast cell lines mRNA expression: CCLE, https://depmap.org/portal/download/all/