Fabian Gröger* · Shuo Wen* · Huyen Le · Maria Brbić
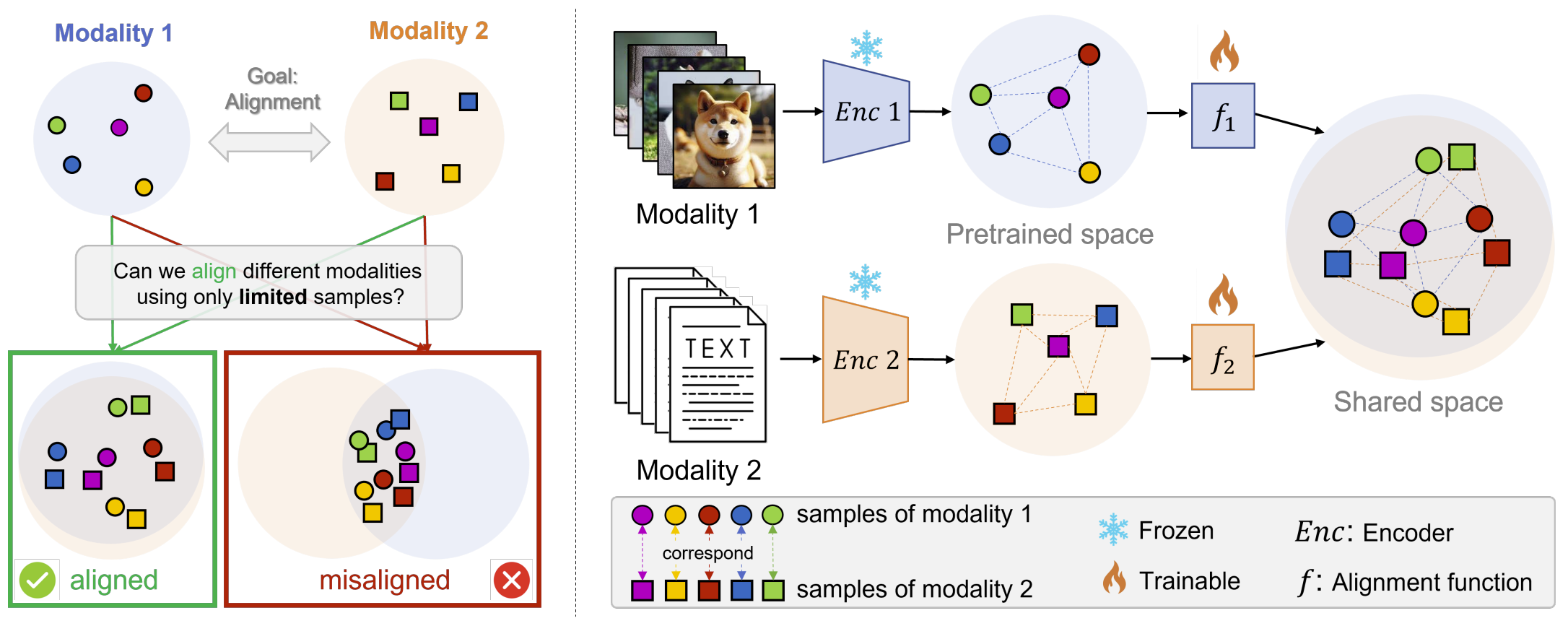
Multimodal models have demonstrated powerful capabilities in complex tasks requiring multimodal alignment, including zero-shot classification and cross-modal retrieval. However, existing models typically rely on millions of paired multimodal samples, which are prohibitively expensive or infeasible to obtain in many domains.
In this work, we explore the feasibility of building multimodal models with limited amounts of paired data by aligning pretrained unimodal foundation models. We show that high-quality alignment is possible with as few as tens of thousands of paired samples — less than 1% of the data typically used in the field.
- STRUCTURE Regularization: An effective technique that preserves the neighborhood geometry of the latent space of unimodal encoders
- Layer Selection: Demonstration that aligning last layers is often suboptimal, with benefits from aligning layers with highest representational similarity
- Strong Results: 51.6% average relative improvement in classification and 91.8% in retrieval tasks across 24 benchmarks
- Broad Applicability: Can be readily incorporated into existing alignment methods
- Python 3.8+
- PyTorch 2.1.2+
- CUDA 11.8+ (for GPU support)
- Clone the repository:
git clone https://github.com/mlbio-epfl/STRUCTURE.git
cd STRUCTURE- Install dependencies:
pip install -r requirements.txtPrepare your datasets using the provided scripts. For example, for COCO:
# COCO dataset will be downloaded automatically
# Place in ./data/coco/For other datasets, see the src/dataset_preparation/ directory for preparation scripts.
Train an alignment model with limited data:
python src/train_subsampled_alignment.py --config_path configs/losses_lin/clip_base_best.yamlTrain with full alignment:
python src/train_alignment.py --config_path configs/losses_lin/clip_base_best.yamlExtract features:
python src/extract_features.py --config_path configs/default.yamlMeasure alignment quality:
python src/measure_alignment.py --config_path configs/metrics/clip_mutual_knn_rice.yamlThe repository uses YAML configuration files located in configs/:
configs/default.yaml- Base configurationconfigs/losses_lin/- Linear alignment layer configurationsconfigs/losses_mlp/- MLP alignment layer configurationsconfigs/ablations/- Ablation study configurationsconfigs/csa/- CSA configurationsconfigs/metrics/- Alignment metrics configurations
representation-alignment/
├── configs/ # Configuration files
├── src/
│ ├── alignment/ # Alignment layer implementations
│ ├── trainers/ # Training logic
│ ├── models/ # Model implementations
│ ├── loss/ # Loss functions
│ ├── evaluation/ # Evaluation metrics
│ ├── dataset_preparation/ # Dataset preparation scripts
│ └── utils/ # Utility functions
├── data/ # Dataset directory (created during setup)
└── requirements.txt # Python dependencies
If you find this work useful, please cite our paper:
@inproceedings{groger2025structure,
title={With Limited Data for Multimodal Alignment, Let the {STRUCTURE} Guide You},
author={Gr{\"o}ger, Fabian and Wen, Shuo and Le, Huyen and Brbic, Maria},
booktitle={The Thirty-ninth Annual Conference on Neural Information Processing Systems},
year={2025},
url={https://openreview.net/forum?id=IkvQqD7hk3}
}