-
Notifications
You must be signed in to change notification settings - Fork 4
Add derotation blogpost #163
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Merged
Merged
Changes from 3 commits
Commits
Show all changes
9 commits
Select commit
Hold shift + click to select a range
d3e9955
Add blogpost
lauraporta dbf129a
Linkcheck ignore as biorxiv is very slow
lauraporta 2d9557e
Revert "Linkcheck ignore as biorxiv is very slow"
lauraporta 9af94d5
Improve blogpost also thanks to suggestions
lauraporta ce91918
Include figures and suggested modifications
lauraporta 36d60c6
Exclude full text links for linkcheck error
lauraporta 607d44d
Linkcheck fix
lauraporta 5635429
Improve styling - css center images - ensure ngs remain transparent i…
lauraporta 6847ce2
More links to be added to linkcheck ignore :(
lauraporta File filter
Filter by extension
Conversations
Failed to load comments.
Loading
Jump to
Jump to file
Failed to load files.
Loading
Diff view
Diff view
There are no files selected for viewing
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,83 @@ | ||
| :blogpost: true | ||
| :date: July 1, 2025 | ||
| :author: Laura Porta | ||
| :location: London, UK | ||
| :category: Blog | ||
| :language: English | ||
| :image: 1 | ||
|
|
||
| # `derotation`: a Python package for correcting motion artifacts in rotating multiphoton movies | ||
|
|
||
| The `derotation` package provides a robust solution for reconstructing multiphoton movies of rotating samples acquired with a line-scanning microscope at high speeds and low frame rates. When imaging a rotating sample, the line-by-line nature of acquisition introduces significant geometric distortions. This package corrects these artifacts to produce clear, stable movies suitable for standard analysis pipelines like Suite2p. | ||
lauraporta marked this conversation as resolved.
Outdated
Show resolved
Hide resolved
|
||
|
|
||
| 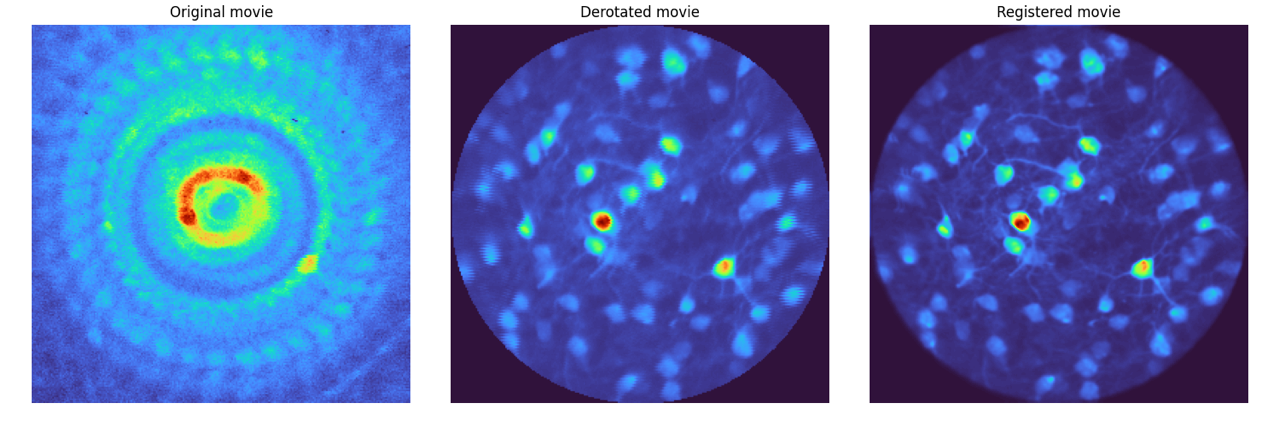 | ||
| > **Left**: The mean image from a raw 3-photon movie of a rotating sample. **Center**: The mean image after processing with `derotation`. **Right**: The mean image after further registration with Suite2p. The improvement in cell definition is evident after derotation. | ||
lauraporta marked this conversation as resolved.
Outdated
Show resolved
Hide resolved
|
||
|
|
||
| --- | ||
|
|
||
| ## Core Functionality: Line-by-Line Correction | ||
|
|
||
| The fundamental principle of the package is **derotation-by-line**. Because a line-scanning microscope acquires an image one horizontal line at a time, a rotating sample causes each line to be captured at a slightly different angle. If the angle of rotation is recorded simultaneously, the `derotation` package can computationally rotate each line back to a common frame of reference, incrementally building a corrected, distortion-free image. | ||
|
|
||
|  | ||
lauraporta marked this conversation as resolved.
Outdated
Show resolved
Hide resolved
|
||
| > This animation demonstrates the line-by-line reconstruction. The distorted frame (left) is corrected one line at a time based on its specific rotation angle to generate the final, stabilized frame (right). | ||
|
|
||
| --- | ||
|
|
||
| ## How to Use `derotation` | ||
|
|
||
| The package can be used in two primary ways, offering a trade-off between granular control and automation. | ||
|
|
||
| ### 1. Low-Level Core Function | ||
| For direct control, users can call the `derotate_an_image_array_line_by_line` function. This requires two main inputs: | ||
| * The raw multiphoton movie (as a NumPy array). | ||
| * The corresponding rotation angle for each captured line. | ||
lauraporta marked this conversation as resolved.
Show resolved
Hide resolved
|
||
|
|
||
| This approach is ideal for users who have already processed their rotation signals and need to apply the core correction algorithm. | ||
|
|
||
| ### 2. High-Level Pipelines | ||
| For an end-to-end workflow, `derotation` provides two pre-built pipeline classes that automate the entire process from raw data to a corrected movie file. | ||
|
|
||
| * **`FullPipeline`**: Designed for experiments with randomized rotations. It automatically parses analog signals, interpolates angles, uses Bayesian optimization to find the center of rotation, and performs the derotation. | ||
| * **`IncrementalPipeline`**: Tailored for continuous, slow rotations. | ||
|
|
||
| Both pipelines generate a corrected TIFF stack, metadata files, and debugging plots. | ||
|
|
||
| --- | ||
|
|
||
| ## Finding the Center of Rotation | ||
|
|
||
| Accurately identifying the **center of rotation** is critical. An incorrect center will result in residual motion, where stationary objects appear to trace circles in the final movie. | ||
|
|
||
| 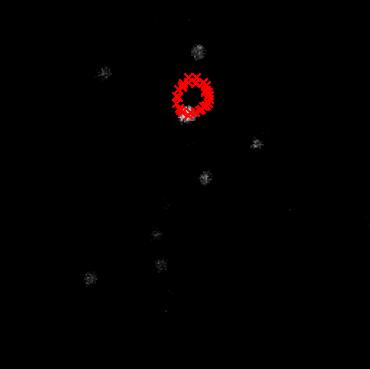 | ||
| > When the center of rotation is miscalculated, a stationary cell's center (red crosses) will appear to move in a circular path across different rotation angles. | ||
|
|
||
| The package implements two methods to find this center: | ||
|
|
||
| 1. **Bayesian Optimization**: Used by the `FullPipeline`, this method is computationally intensive but robust. It iteratively tests potential center coordinates to find the one that minimizes motion in the corrected movie. | ||
lauraporta marked this conversation as resolved.
Show resolved
Hide resolved
|
||
| 2. **Ellipse Fitting**: The `IncrementalPipeline` tracks the path of a bright, stable object (like a cell) across frames. In a physical rotating system, this path would likely form an ellipse. The center of the fitted ellipse is an accurate estimate of the true center of rotation. | ||
|
|
||
| --- | ||
|
|
||
| ## Verification and Synthetic Data | ||
|
|
||
| To ensure the quality of the correction and validate the algorithms, `derotation` includes tools for generating and processing synthetic data. | ||
|
|
||
| * **`Rotator` Class**: This class can take a static image and apply a virtual line-by-line rotation, simulating the distortion from a real microscope. | ||
| * **`SyntheticData` Class**: This builds on `Rotator` to create complete, simulated datasets, including fake cell images and corresponding rotation signals. | ||
|
|
||
| 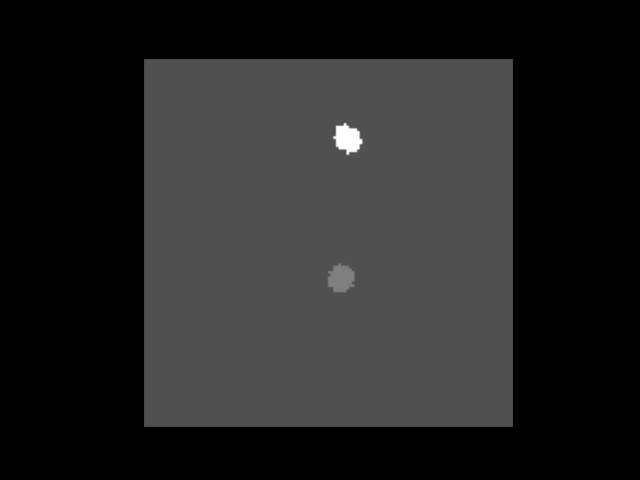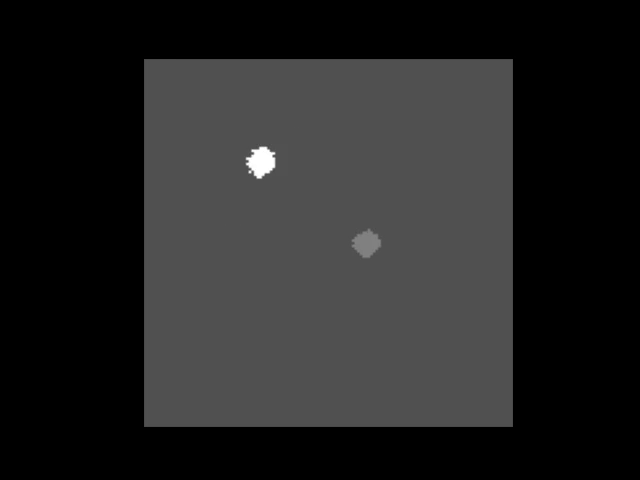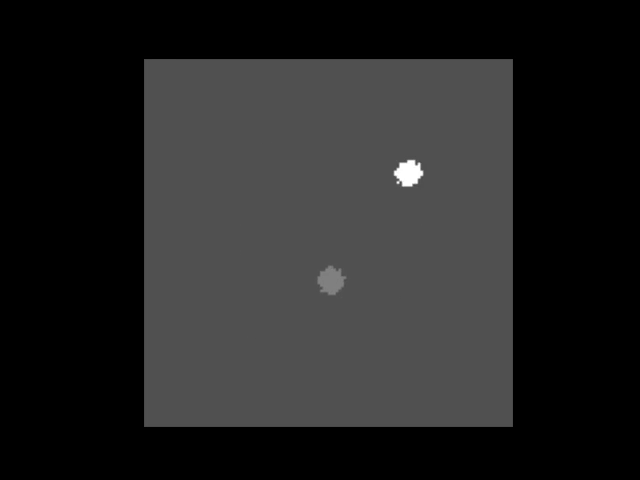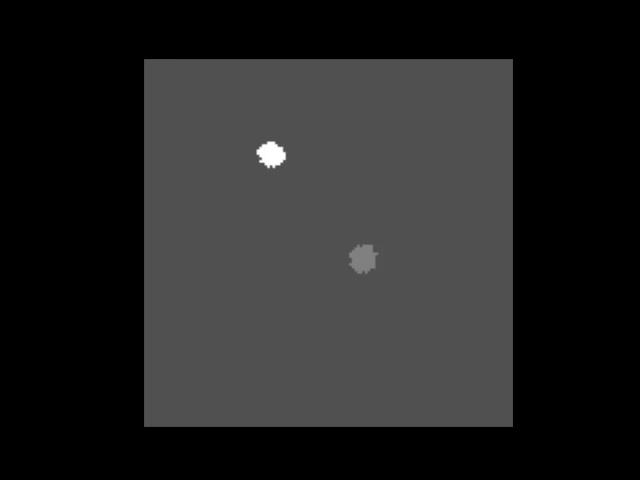 | ||
| > An animation showing a synthetic dataset of two "cells" being rotated with line-scanning artifacts. | ||
|
|
||
| These tools are invaluable for testing and can even simulate complex cases like **out-of-plane rotations**, where the axis of rotation is tilted relative to the imaging plane. In such cases, the package can fit an ellipse to the object's trajectory to calculate the 3D rotation geometry and apply a more sophisticated correction. | ||
|
|
||
| --- | ||
|
|
||
| ## Get Involved | ||
|
|
||
| `derotation` is an open-source project in active development. Community contributions are welcome. | ||
|
|
||
| * **Report Bugs or Request Features**: [Open an issue on GitHub](https://github.com/neuroinformatics-unit/derotation/issues) | ||
lauraporta marked this conversation as resolved.
Outdated
Show resolved
Hide resolved
|
||
| * **Discuss Development**: [Join the Zulip chat](https://neuroinformatics.zulipchat.com/#narrow/channel/495735-Derotation) | ||
|
|
||
| This project is sponsored by the Margrie Lab at the Sainsbury Wellcome Centre. | ||
Add this suggestion to a batch that can be applied as a single commit.
This suggestion is invalid because no changes were made to the code.
Suggestions cannot be applied while the pull request is closed.
Suggestions cannot be applied while viewing a subset of changes.
Only one suggestion per line can be applied in a batch.
Add this suggestion to a batch that can be applied as a single commit.
Applying suggestions on deleted lines is not supported.
You must change the existing code in this line in order to create a valid suggestion.
Outdated suggestions cannot be applied.
This suggestion has been applied or marked resolved.
Suggestions cannot be applied from pending reviews.
Suggestions cannot be applied on multi-line comments.
Suggestions cannot be applied while the pull request is queued to merge.
Suggestion cannot be applied right now. Please check back later.
Uh oh!
There was an error while loading. Please reload this page.