-
Notifications
You must be signed in to change notification settings - Fork 0
Split readme docs #114
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Split readme docs #114
Changes from 5 commits
6390a78
b2afb2a
1440faf
99c4881
57ad6c9
be76202
b2cb85d
ff7cdef
43a7b7f
822e447
468c9af
fa391a9
71f1f7f
11403ef
56fa147
File filter
Filter by extension
Conversations
Jump to
Diff view
Diff view
There are no files selected for viewing
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -27,6 +27,9 @@ jobs: | |
| run: | | ||
| python -m pip install --upgrade pip | ||
| pip install -e .[docs] | ||
| - name: Split README into sections | ||
| run: | | ||
| python docs/split_readme.py | ||
|
||
| - name: Build references | ||
| run: | | ||
| cd docs | ||
|
|
||
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,6 +1,3 @@ | ||
| # Vuegen Case Study - View HTML Report | ||
| # Earth Microbiome Project Case Study - HTML and Streamlit Example Reports | ||
|
|
||
| The stable version of the current `html` report generated in the example notebook | ||
| using `vuegen` can be viewed at | ||
|
|
||
| [multiomics-analytics-group.github.io/vuegen/](https://multiomics-analytics-group.github.io/vuegen/) | ||
| The Earth Microbiome Project case study generated in the example notebook using `vuegen` is available online as [HTML](https://multiomics-analytics-group.github.io/vuegen/) and [Streamlit](https://earth-microbiome-vuegen-demo.streamlit.app/) reports. |
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,17 @@ | ||
| ## About the project | ||
|
|
||
| **VueGen** is a tool that automates the creation of **reports** from bioinformatics outputs, allowing researchers with minimal coding experience to communicate their results effectively. With VueGen, users can produce reports by simply specifying a directory containing output files, such as plots, tables, networks, Markdown text, HTML components, and API calls, along with the report format. Supported formats include **documents** (PDF, HTML, DOCX, ODT), **presentations** (PPTX, Reveal.js), **Jupyter notebooks**, and [Streamlit](https://streamlit.io/) **web applications**. | ||
|
|
||
| A YAML configuration file is generated from the directory to define the structure of the report. Users can customize the report by modifying the configuration file, or they can create their own configuration file instead of passing a directory as input. The configuration file specifies the structure of the report, including sections, subsections, and various components such as plots, dataframes, markdown, html, and API calls. | ||
|
|
||
| An overview of the VueGen workflow is shown in the figure below: | ||
|
|
||
| 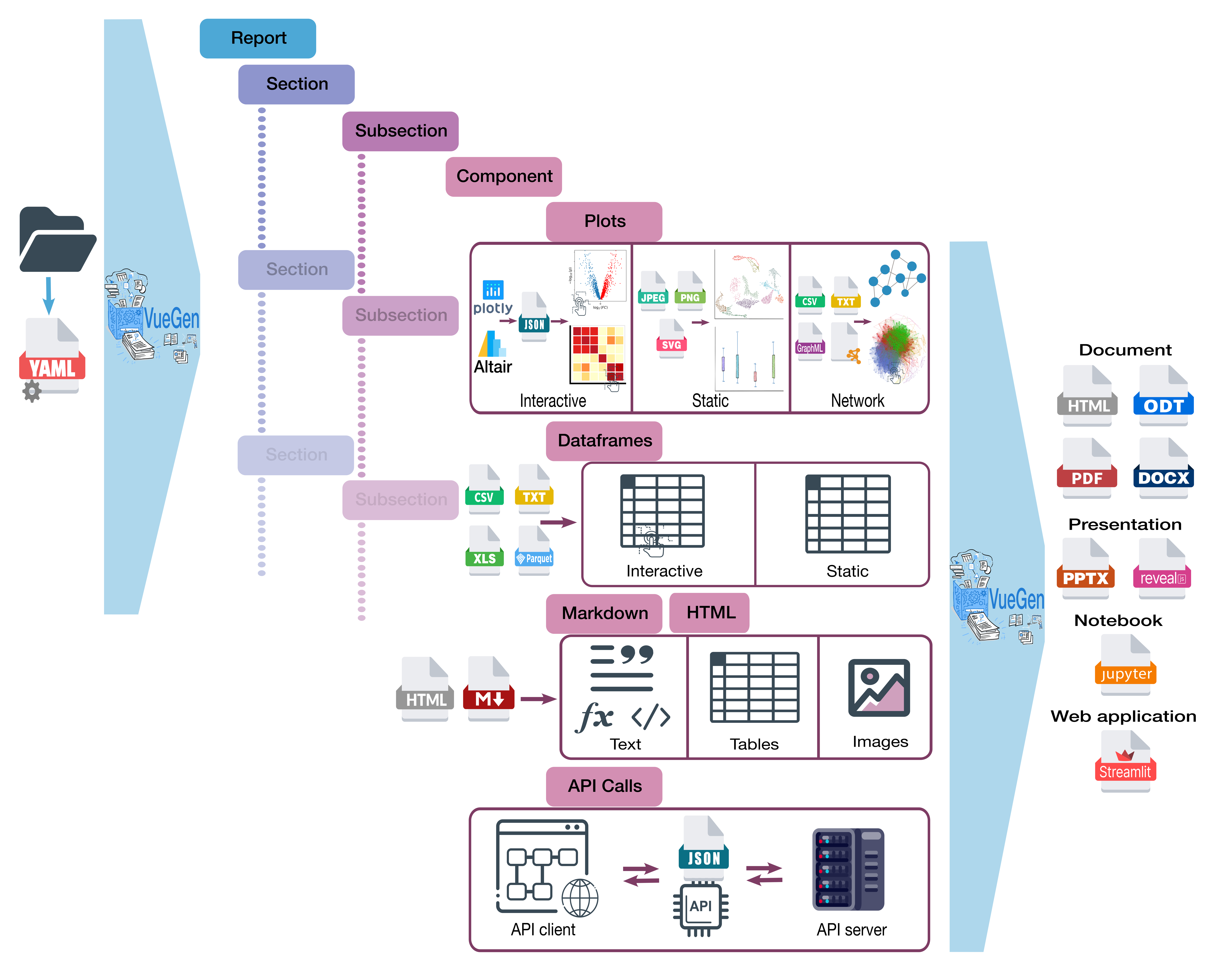 | ||
|
|
||
| Also, the class diagram for the project is presented below to illustrate the architecture and relationships between classes: | ||
|
|
||
| 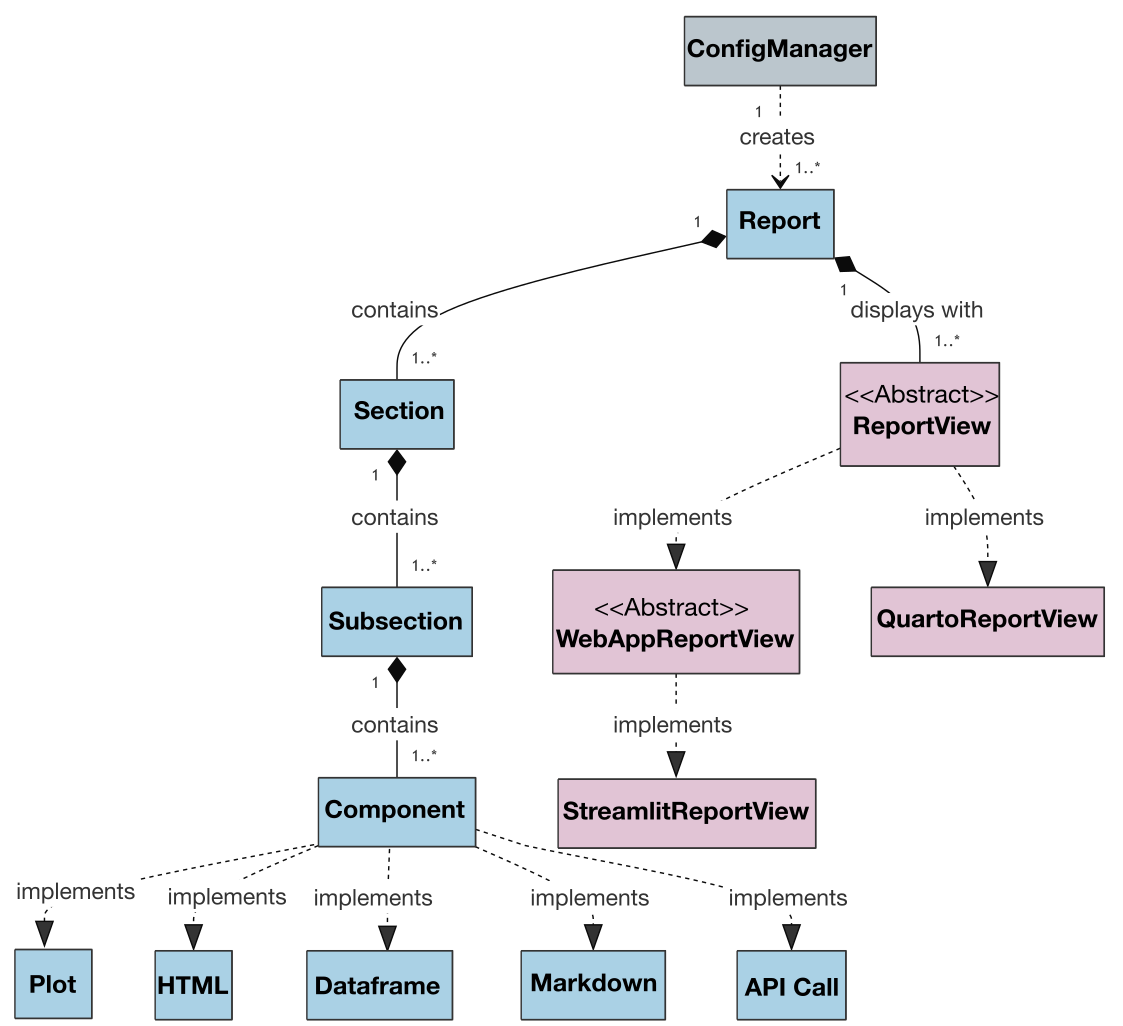 | ||
|
|
||
| An extended version of the class diagram with attributes and methods is available [here](https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_classdiagram_withattmeth.pdf). | ||
|
|
||
| The VueGen documentation is available at [vuegen.readthedocs.io](https://vuegen.readthedocs.io/), where you can find detailed information of the package’s classes and functions, installation and execution instructions, and case studies to demonstrate its functionality. |
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,58 @@ | ||
| ## Case studies | ||
|
||
|
|
||
| VueGen’s functionality is demonstrated through two case studies: | ||
|
|
||
| **1. Predefined Directory** | ||
|
||
|
|
||
| This introductory case study uses a predefined directory with plots, dataframes, Markdown, and HTML components. Users can generate reports in different formats and modify the configuration file to customize the report structure. | ||
|
|
||
| 🔗 [](https://colab.research.google.com/github/Multiomics-Analytics-Group/vuegen/blob/main/docs/vuegen_basic_case_study.ipynb) | ||
|
|
||
| :::{NOTE} | ||
| The [configuration file](https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/docs/example_config_files/Basic_example_vuegen_demo_notebook_config.yaml) is available in the `docs/example_config_files` folder, and the [directory](https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/docs/example_data/Basic_example_vuegen_demo_notebook) with example data is in the `docs/example_data` folder. | ||
| ::: | ||
|
|
||
|
|
||
| **2. Earth Microbiome Project Data** | ||
|
|
||
| This advanced case study demonstrates the application of VueGen in a real-world scenario using data from the [Earth Microbiome Project (EMP)](https://earthmicrobiome.org/). The EMP is an initiative to characterize global microbial taxonomic and functional diversity. The notebook process the EMP data, create plots, dataframes, and other components, and organize outputs within a directory to produce reports. Report content and structure can be adapted by modifying the configuration file. Each report consists of sections on exploratory data analysis, metagenomics, and network analysis. | ||
|
|
||
| 🔗 [](https://colab.research.google.com/github/Multiomics-Analytics-Group/vuegen/blob/main/docs/vuegen_case_study_earth_microbiome.ipynb) | ||
|
|
||
| :::{NOTE} | ||
| The EMP case study is available online as [HTML](https://multiomics-analytics-group.github.io/vuegen/) and [Streamlit](https://earth-microbiome-vuegen-demo.streamlit.app/) reports. | ||
| The [configuration file](https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/docs/example_config_files/Earth_microbiome_vuegen_demo_notebook_config) is available in the `docs/example_config_files` folder, and the [directory](https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/docs/example_data/Earth_microbiome_vuegen_demo_notebook) with example data is in the `docs/example_data` folder. | ||
| ::: | ||
|
|
||
|
|
||
| **3. ChatBot Component** | ||
|
|
||
| This case study highlights VueGen’s capability to embed a chatbot component into a report subsection, | ||
| enabling interactive conversations inside the report. | ||
|
|
||
| Two API modes are supported: | ||
|
|
||
| - **Ollama-style streaming chat completion** | ||
| If a `model` parameter is specified in the config file, VueGen assumes the chatbot is using Ollama’s [/api/chat endpoint](https://github.com/ollama/ollama/blob/main/docs/api.md#generate-a-chat-completion). | ||
| Messages are handled as chat history, and the assistant responses are streamed in real time for a smooth and responsive experience. | ||
| This mode supports LLMs such as `llama3`, `deepsek`, or `mistral`. | ||
|
|
||
| :::{TIP} | ||
| See [Ollama’s website](https://ollama.com/) for more details. | ||
| ::: | ||
|
|
||
|
|
||
| - **Standard prompt-response API** | ||
| If no `model` is provided, VueGen uses a simpler prompt-response flow. | ||
| A single prompt is sent to an endpoint, and a structured JSON object is expected in return. | ||
| Currently, the response can include: | ||
| - `text`: the main textual reply | ||
| - `links`: a list of source URLs (optional) | ||
| - `HTML content`: an HTML snippet with a Pyvis network visualization (optional) | ||
|
|
||
| This response structure is currently customized for an internal knowledge graph assistant, but VueGen is being actively developed | ||
| to support more flexible and general-purpose response formats in future releases. | ||
|
|
||
| :::{NOTE} | ||
| You can see a [configuration file example](https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/docs/example_config_files/Chatbot_example_config.yaml) for the chatbot component in the `docs/example_config_files` folder. | ||
| ::: | ||
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,22 @@ | ||
| ## Citation | ||
|
||
|
|
||
| If you use VueGen in your research or publications, please cite it as follows: | ||
|
|
||
| **APA:** | ||
|
|
||
| Ayala-Ruano, S., Webel, H., & Santos, A. (2025). _VueGen: Automating the generation of scientific reports_. bioRxiv. https://doi.org/10.1101/2025.03.05.641152 | ||
|
|
||
| **BibTeX:** | ||
|
|
||
| ```bibtex | ||
| @article{Ayala-Ruano2025VueGen, | ||
| author = {Ayala-Ruano, Sebastian and Webel, Henry and Santos, Alberto}, | ||
| title = {VueGen: Automating the generation of scientific reports}, | ||
| journal = {bioRxiv}, | ||
| year = {2025}, | ||
| doi = {10.1101/2025.03.05.641152}, | ||
| publisher = {Cold Spring Harbor Laboratory}, | ||
| url = {https://www.biorxiv.org/content/10.1101/2025.03.05.641152}, | ||
| eprint = {https://www.biorxiv.org/content/10.1101/2025.03.05.641152.full.pdf} | ||
| } | ||
| ``` | ||
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,5 @@ | ||
| ## Credits and acknowledgements | ||
|
|
||
| - VueGen was developed by the [Multiomics Network Analytics Group (MoNA)](https://multiomics-analytics-group.github.io/) at the [Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain)](https://www.biosustain.dtu.dk/). | ||
| - VueGen relies on the work of numerous open-source projects like [Streamlit](streamlit), [Quarto](https://quarto.org/), and others. A big thank you to their authors for making this possible! | ||
| - The vuegen logo was designed based on an image created by [Scriberia](https://www.scriberia.co.uk/) for The [Turing Way Community](https://github.com/the-turing-way/the-turing-way), which is shared under a CC-BY licence. The original image can be found at [Zenodo](https://zenodo.org/records/3695300). |
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,77 @@ | ||
| ## Execution | ||
|
|
||
|
|
||
| :::{IMPORTANT} | ||
| Here we use the `Earth_microbiome_vuegen_demo_notebook` [directory](https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/docs/example_data/Earth_microbiome_vuegen_demo_notebook) and the `Earth_microbiome_vuegen_demo_notebook.yaml` [configuration file](https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/docs/example_config_files/Earth_microbiome_vuegen_demo_notebook_config) as examples, which are available in the `docs/example_data` and `docs/example_config_files` folders, respectively. Make sure to clone this reposiotry to access these contents, or use your own directory and configuration file. | ||
| ::: | ||
|
|
||
|
|
||
| Run VueGen using a directory with the following command: | ||
|
|
||
| ```bash | ||
| vuegen --directory docs/example_data/Earth_microbiome_vuegen_demo_notebook --report_type streamlit | ||
| ``` | ||
|
|
||
| :::{NOTE} | ||
| By default, the `streamlit_autorun` argument is set to False, but you can use it in case you want to automatically run the streamlit app. | ||
| ::: | ||
|
|
||
|
|
||
| ### Folder structure | ||
|
|
||
| Your input directory must follow a **nested folder structure**, where first-level folders are treated as **sections** and second-level folders as **subsections**, containing the components (plots, tables, networks, Markdown text, and HTML files). | ||
|
|
||
| Here is an example layout: | ||
| ``` | ||
| report_folder/ | ||
| ├── section1/ | ||
| │ └── subsection1/ | ||
| │ ├── table.csv | ||
| │ ├── image1.png | ||
| │ └── chart.json | ||
| ├── section2/ | ||
| │ ├── subsection1/ | ||
| │ │ ├── summary_table.xls | ||
| │ │ └── network_plot.graphml | ||
| │ └── subsection2/ | ||
| │ ├── report.html | ||
| │ └── summary.md | ||
| ``` | ||
|
|
||
| :::{WARNING} | ||
| VueGen currently requires each section to contain at least one subsection folder. Defining only sections (with no subsections) or using deeper nesting levels (i.e., sub-subsections) will result in errors. In upcoming releases, we plan to support more flexible directory structures. | ||
| ::: | ||
|
|
||
|
|
||
| The titles for sections, subsections, and components are extracted from the corresponding folder and file names, and afterward, users can add descriptions, captions, and other details to the configuration file. Component types are inferred from the file extensions and names. | ||
| The order of sections, subsections, and components can be defined using numerical suffixes in folder and file names. | ||
|
|
||
| It's also possible to provide a configuration file instead of a directory: | ||
|
|
||
| ```bash | ||
| vuegen --config docs/example_config_files/Earth_microbiome_vuegen_demo_notebook.yaml --report_type streamlit | ||
| ``` | ||
|
|
||
| If a configuration file is given, users can specify titles and descriptions for sections and subsections, as well as component paths and required attributes, such as file format and delimiter for dataframes, plot types, and other details. | ||
|
|
||
| The current report types supported by VueGen are: | ||
|
|
||
| - Streamlit | ||
| - HTML | ||
| - DOCX | ||
| - ODT | ||
| - Reveal.js | ||
| - PPTX | ||
| - Jupyter | ||
|
|
||
| ### Running VueGen with Docker | ||
|
|
||
| Instead of installing VueGen locally, you can run it directly from a Docker container with the following command: | ||
|
|
||
| ```bash | ||
| docker run --rm \ | ||
| -v "$(pwd)/docs/example_data/Earth_microbiome_vuegen_demo_notebook:/home/appuser/Earth_microbiome_vuegen_demo_notebook" \ | ||
| -v "$(pwd)/output_docker:/home/appuser/streamlit_report" \ | ||
| quay.io/dtu_biosustain_dsp/vuegen:v0.3.2-docker --directory /home/appuser/Earth_microbiome_vuegen_demo_notebook --report_type streamlit | ||
| ``` |
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,57 @@ | ||
| ## GUI | ||
|
|
||
| ### Local GUI | ||
|
|
||
| We have a simple GUI for VueGen that can be run locally or through a standalone executable. | ||
| For now you will need to have a copy of this repository. | ||
|
|
||
| ```bash | ||
| pip install '.[gui]' | ||
| cd gui | ||
| python app.py | ||
| ``` | ||
|
|
||
| ### Bundled GUI | ||
|
|
||
| The **bundle GUI** with the VueGen package is available under the | ||
| [latest releases](https://github.com/Multiomics-Analytics-Group/vuegen/releases/latest). | ||
| You will need to unzip the file and run `vuegen_gui` in the unpacked main folder. | ||
| Most dependencies are included into the bundle using PyInstaller. | ||
|
|
||
| Streamlit works out of the box as a purely Python based package. For `html` creation you will have to | ||
| have a Python 3.12 installation with the `jupyter` package installed as `quarto` needs to start | ||
| a kernel for execution. This is also true if you install `quarto` globally on your machine. | ||
|
|
||
| We recommend using miniforge to install Python and the conda package manager: | ||
|
|
||
| - [conda-forge.org/download/](https://conda-forge.org/download/) | ||
|
|
||
| We continous our example assuming you have installed the `miniforge` distribution for your | ||
| machine (MacOS with arm64/ apple silicon or x86_64/ intel or Windows x86_64). Also download | ||
| the [latest `vuegen_gui` bundle](https://github.com/Multiomics-Analytics-Group/vuegen/releases/latest) | ||
| from the releases page according to your operating system. | ||
|
|
||
| ```bash | ||
| conda create -n vuegen_gui -c conda-forge python=3.12 jupyter | ||
| conda info -e # find environment location | ||
| ``` | ||
|
|
||
| Find the vuegen_gui path for your local `user`. | ||
|
|
||
| On **MacOS** you need to add a `bin` to the path: | ||
|
|
||
| ```bash | ||
| /Users/user/miniforge3/envs/vuegen_gui/bin | ||
| ``` | ||
|
|
||
| On **Windows** you can use the path as displayed by `conda info -e`: | ||
|
|
||
| > Note: On Windows a base installation of miniforge with `jupyter` might work as well as | ||
| > the app can see your entire Path which is not the case on MacOS. | ||
|
|
||
| ```bash | ||
| C:\Users\user\miniforge3\envs\vuegen_gui | ||
| ``` | ||
|
|
||
| More information regarding the app and builds can be found in the | ||
| [GUI README](https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/gui/README.md). |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Is the
sphinx-apidoccommand here also needed considering it's also done on the sphinx conf.py?There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This action just tests that the execution works. As the
index.mdexpects some file under references, it is needed here.