A metric for the evaluation of single-cell query-to-reference mappings
Please refer to the documentation, in particular, the API documentation for detailed package documentation. For reproduction of the results in the paper, check out the mapQC reproducibility repository.
Below a few notes on how and when to use mapQC:
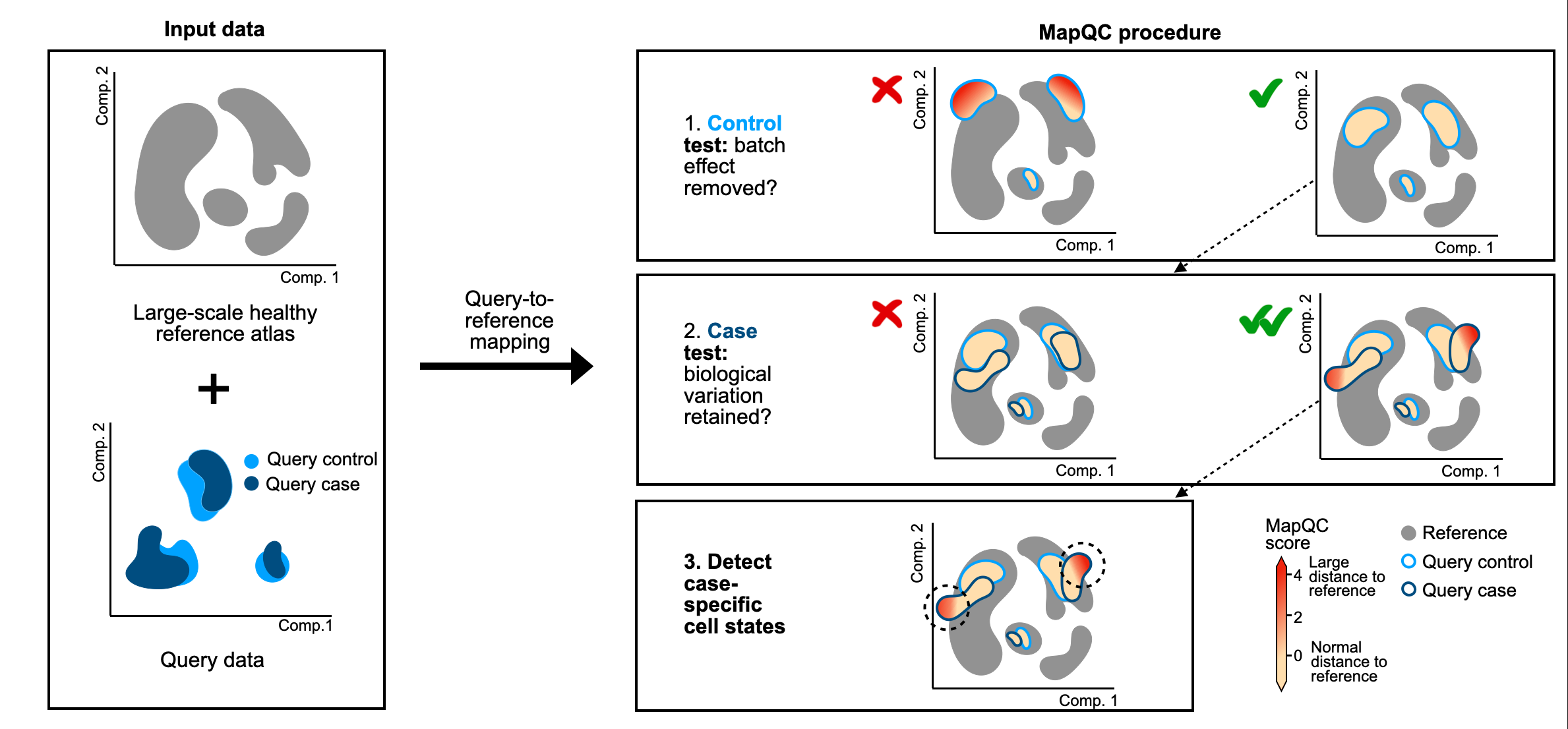
MapQC evaluates the quality of a query-to-reference mapping, and outputs a cell-level mapQC score for every query cell. MapQC scores higher than 2 indicate a large distance of the query cell to the reference. Given a healthy/control reference, we expect query controls to have low mapQC scores, and query case/disease cells to have higher mapQC scores in the case of case-specific cellular phenotypes. You can thus use mapQC scores to assess, in a quantitative manner, if your mapping was successful.
Overview of mapQC's workflow
In short, you need one AnnData object, including:
- A large scale reference, including only its healthy/control cells.
- A mapped query dataset, with healthy/control cells (must-have) and case/perturbed cells (if you have them).
- Metadata (query/reference status, study, sample, and optionally clustering and cell type annotations)
- A mapping-derived embedding including both the reference and the query
In the quick-start tutorial notebook we provide a more extensive description of the exact data requirements.
You need to have Python 3.10 or newer installed on your system.
There are several alternative options to install mapQC:
- Install the latest release of
mapqcfrom PyPI:
pip install mapqc- Install the latest development version:
pip install git+https://github.com/theislab/mapqc.git@mainSee the changelog.
I am happy to hear any comments, suggestions, or even bugs that you run into. I would like to make this package run as smoothly as possible! So for any of these, submit an issue on the mapQC GitHub page and I will be glad to help.
Sikkema et al., bioRxiv 2025.